Simple Visualizers¶
Brain Activity Visualizer¶
A 3D scene of the brain activity.
Mouse interaction:
You can change the view by pressing a mouse button and dragging it.
the left button rotates the brain around the center of the screen.
the right button translates the brain.
the middle button and the scroll wheel zoom towards the center of the screen.
Pressing the shift key and the left button has the same effect as the right button.
Pressing the control key will rotate or translate in the model space; while without control key pressed, the rotation happens in the space of the navigator (with center in (0,0,0) ).
The SPACE key will show a top view. The CURSOR Keys will show axis aligned views.
For region level time series the brain is represented by a coarse granularity - each region is represented with only one color. For surface level time series each vertex has an individual measure.
The color coding is determined by the current color scheme. A legend of it is on the right side of the brain view. You can change this color scheme and other viewer parameters from the brain menu in the upper right corner.
From the visualizer toolbar you can pause and resume the activity movie. For region level time series there is a selection component in the toolbar. Use it to show activity only for the selected regions.
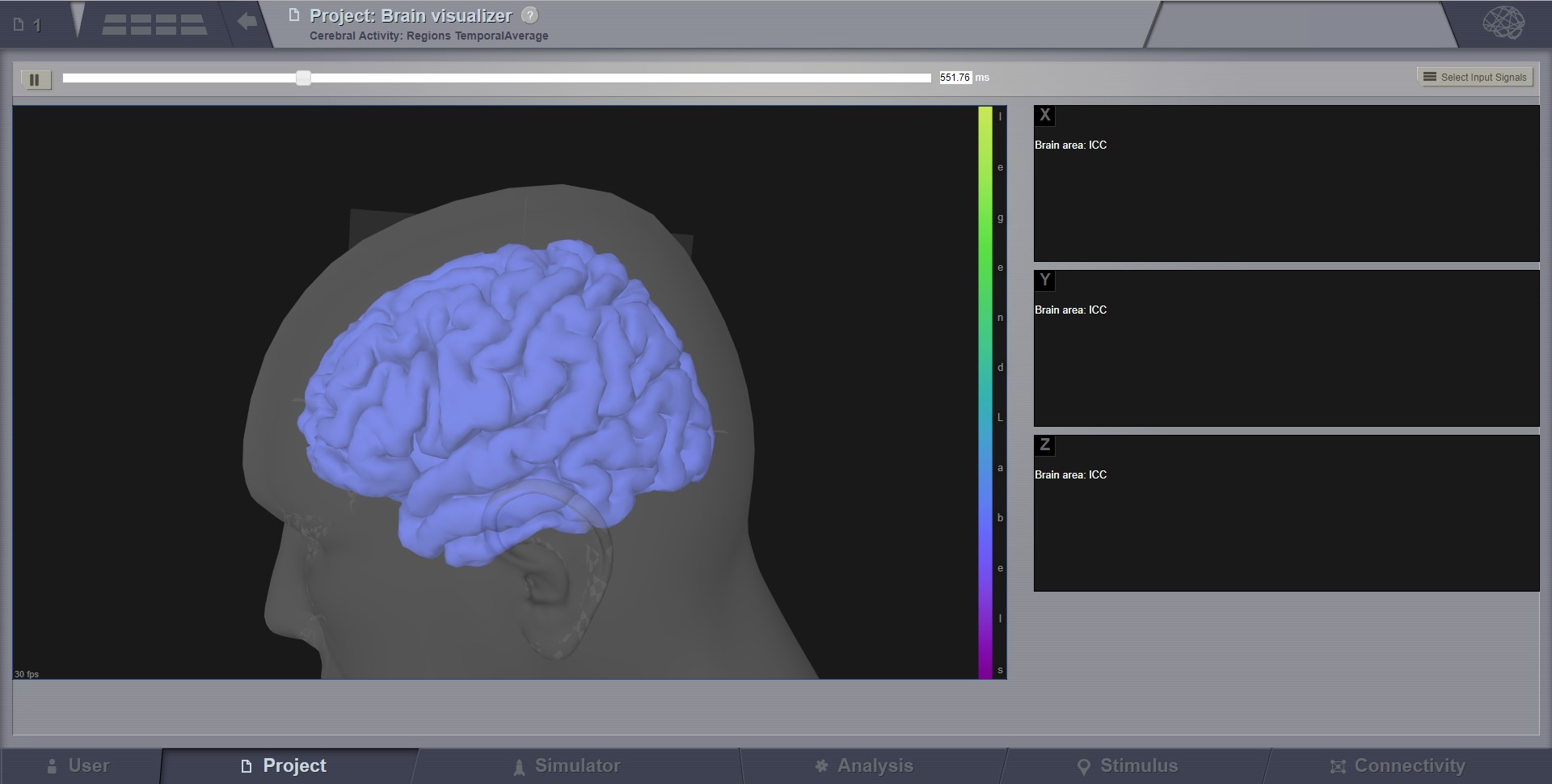
Preview for Brain Activity Visualizer at the region level¶
Time Series Visualizer (svg/d3)¶
In the center area you click and drag to zoom, click once to reset zoom and use the scroll wheel to scroll signals.
The horizontal bottom part is the temporal context. Here the solid line marks the mean across channels, in time. The shaded area marks standard deviation across channels, in time. You Click and drag to select a subset of signals. The selection can be changed again by dragging it. Click outside selection box to cancel and reset view. You can resize the view by dragging blue box in the bottom right corner.
The vertical left part is the signal context. Here solid lines show each signal. Selection works like in the temporal context.
In the brain menu there is a slider you use to change the signal scaling.
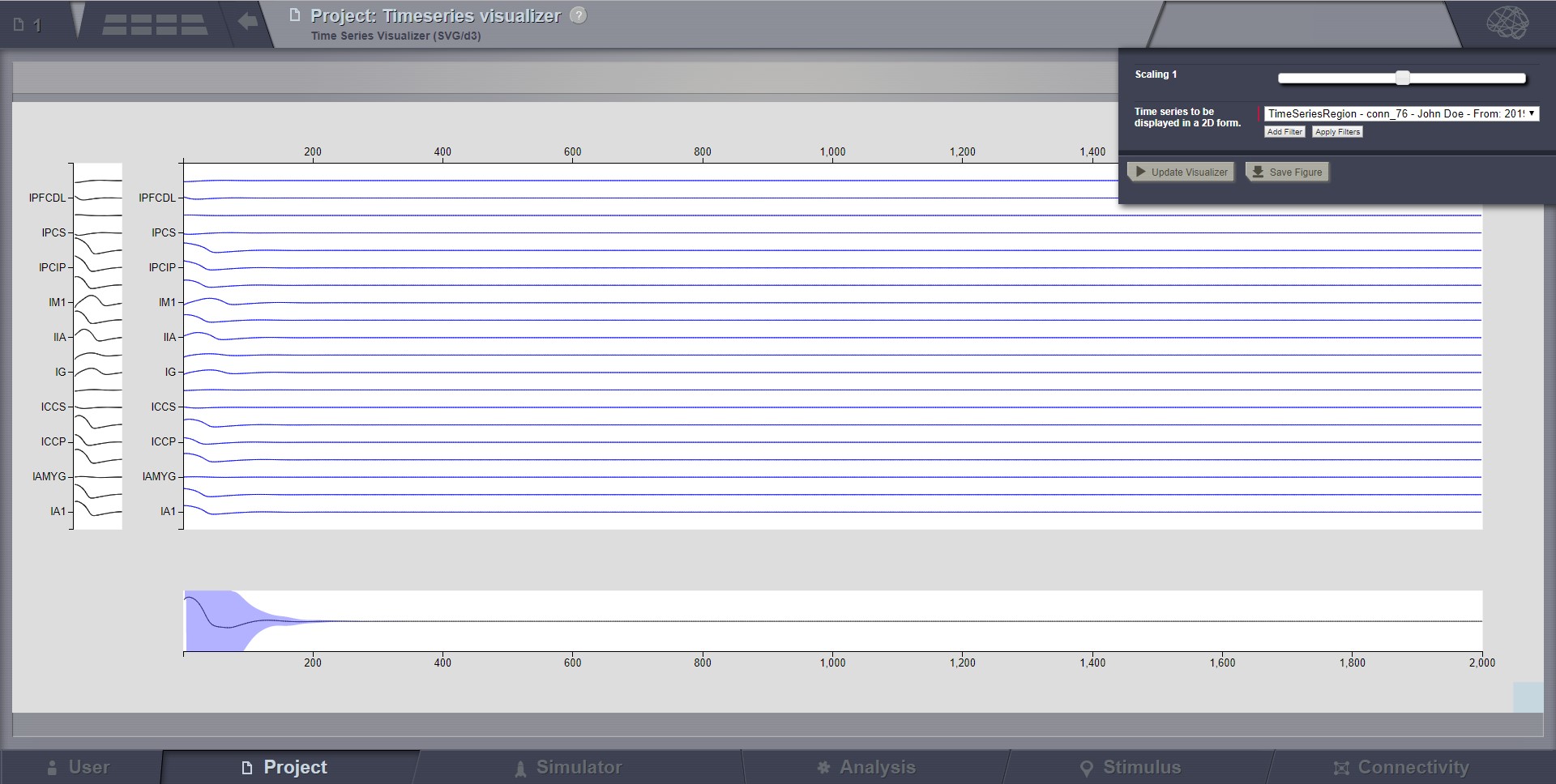
Preview for Time-Series Visualizer (svg/d3)¶
Animated Time Series Visualizer¶
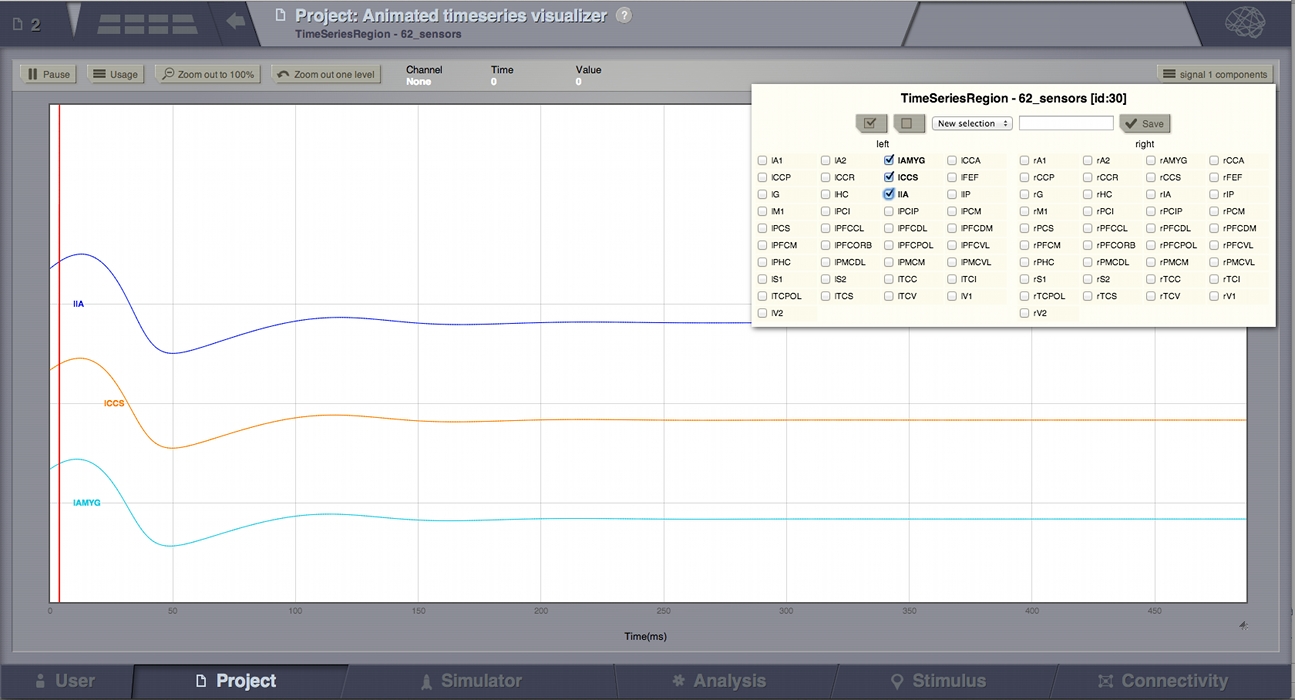
Preview for Animated Time Series Visualizer¶
This is an alternative for the Time Series Visualizer (svg/d3). It is used to display signal lines in 2D.
The label “animated” comes from the red line which will pass the entire signal step by step, at a configurable speed. In single mode, this red-line might not be very useful, but it makes more sense when the same 2D display gets reused in the Dual Visualizers (combined with the 3D display on a surface) where the red-line shows the current step displayed in the 3D movie on the left.
Select zoom area with your mouse (you may do that several times to zoom in further). From the toolbar you can pause resume the activity and zoom out.
This viewer can display multiple time series. On the right side of the toolbar there will be a selection component for each signal source. These selection components determine what signals are shown in the viewer. To select additional time series use the brain menu in the upper left corner. From that menu you can change viewer settings. The page size determines how much data should appear at once in the viewer. The spacing determines the space between the horizontal axis of each signal. Setting it to 0 will plot all signals in the same coordinate system. A side effect of this setting is that as you decrease this axis separation the amplitude of signals is scaled up.
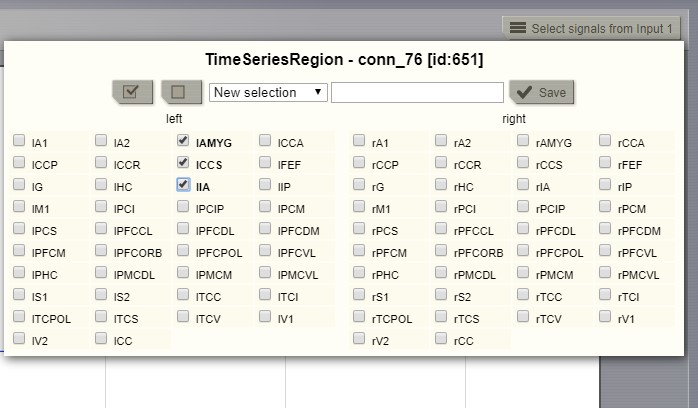
Selecting the “channels” to be displayed (available in several viewers of TVB).¶
Dual Brain Activity Visualizer¶
This visualizer combines the brain activity movie shown in a 3D display on the left, with the explicit channels recording lines on the right. Movie start/stop, speed control, color schema change, channel selection are some of the features available in this visualizer.
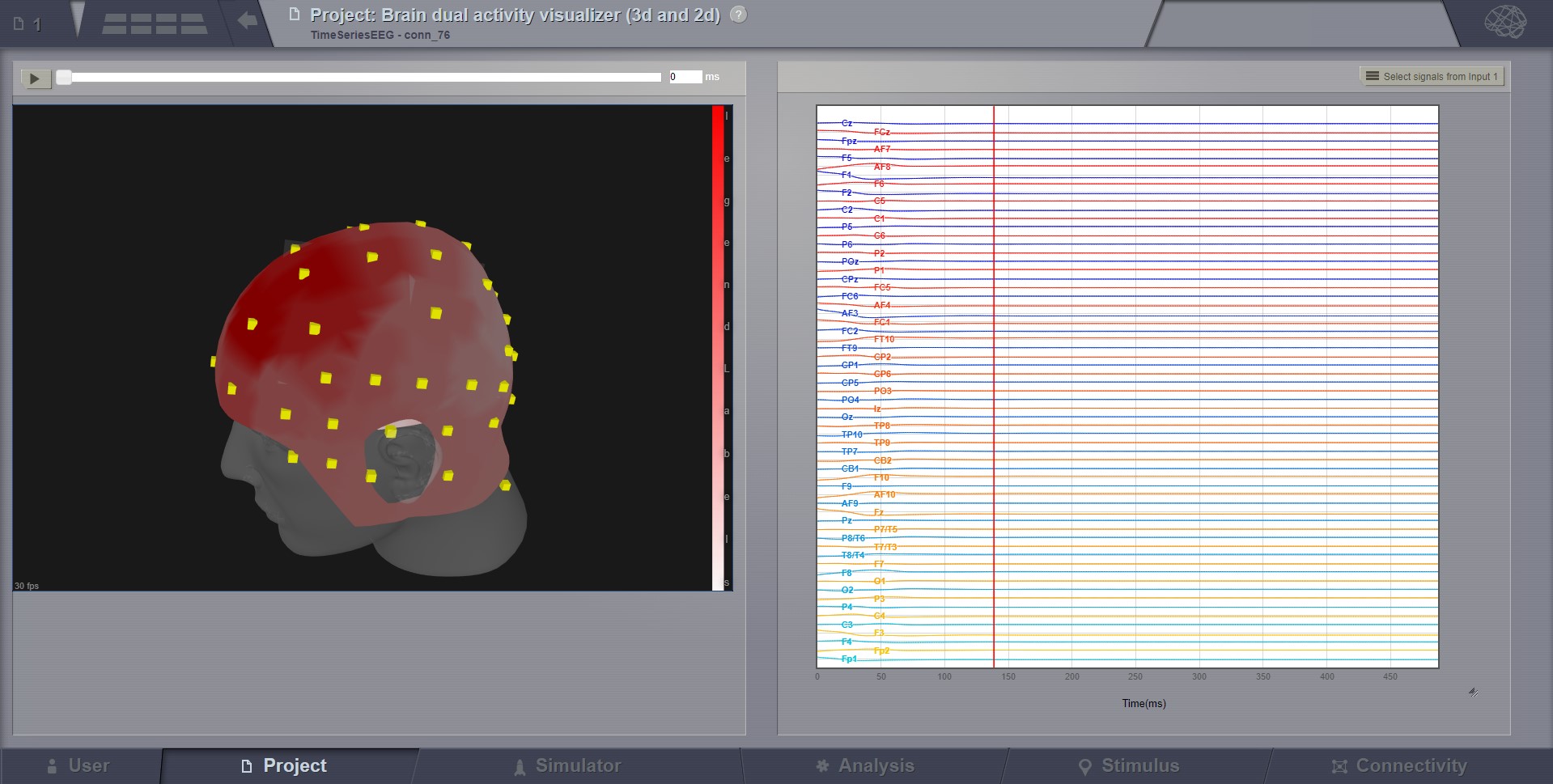
Brain activity with EEG recordings.¶
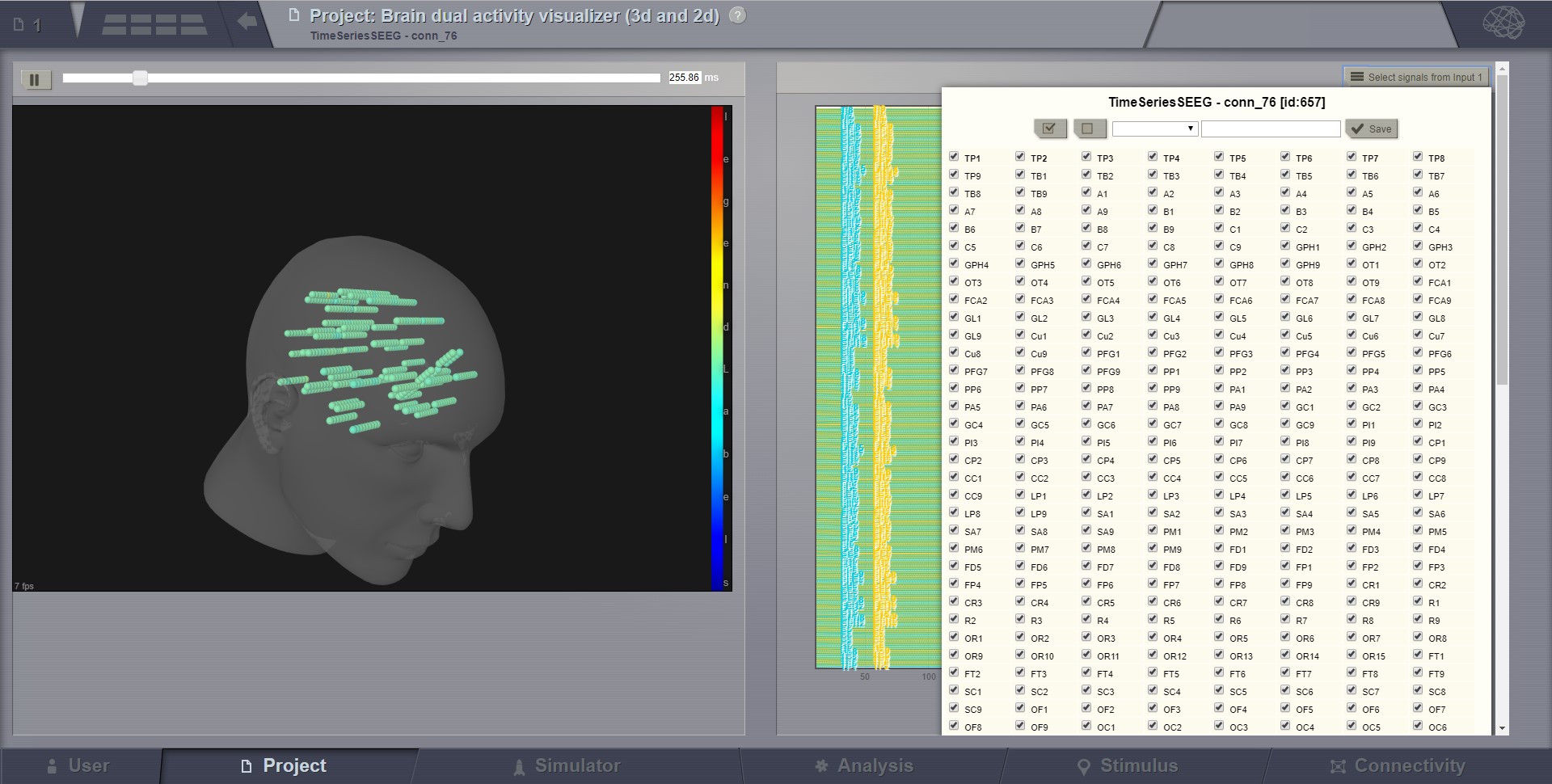
Brain activity with sEEG recordings.¶
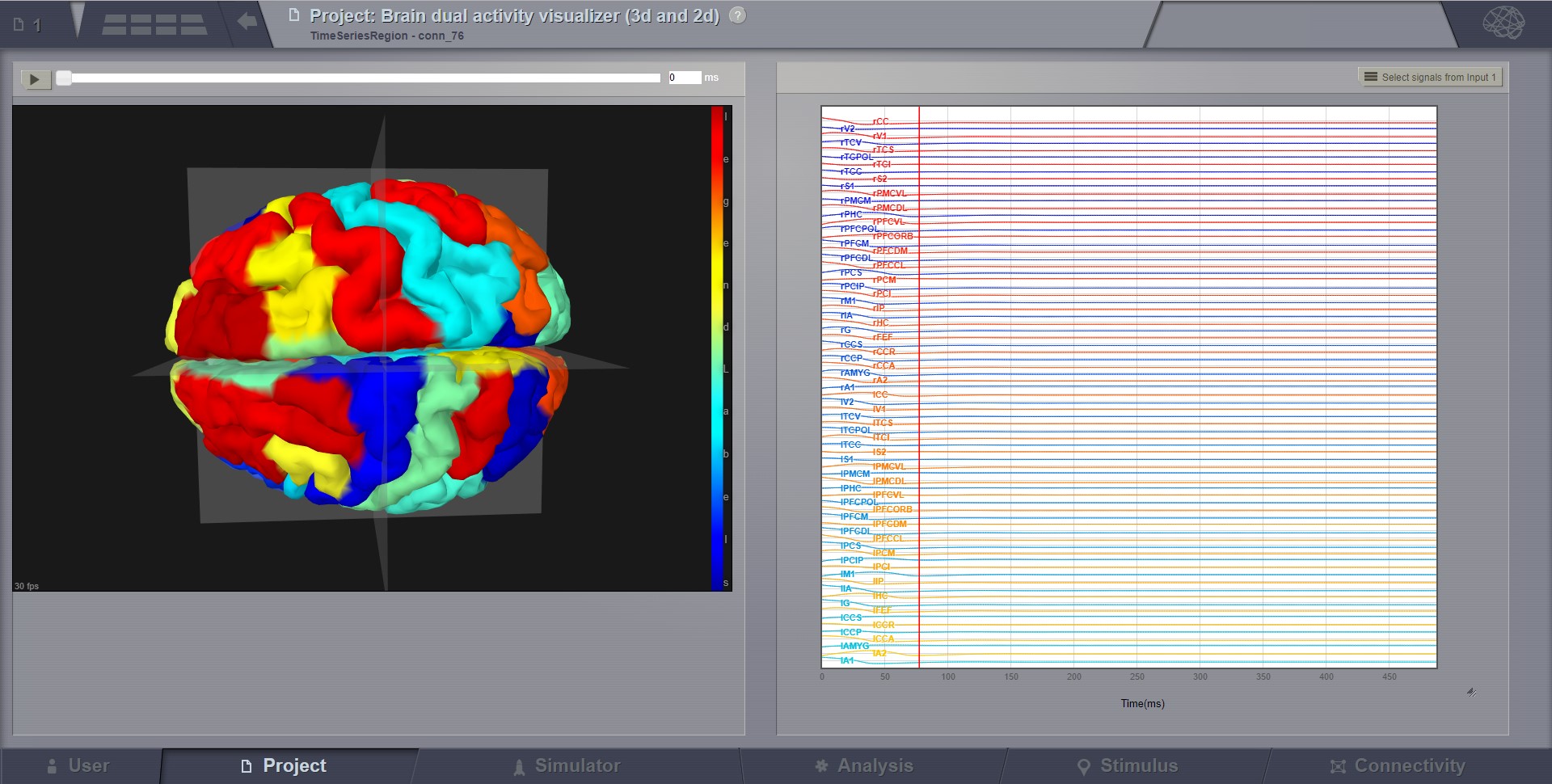
Brain activity with region level activity.¶
Volume Visualizer¶
This family of viewers display volumetric data, usually in an anatomical space. If the data has a time component then on the right side it will display timelines for selected voxels. FMri data is an example of this. A structural mri volume may be used as a background.
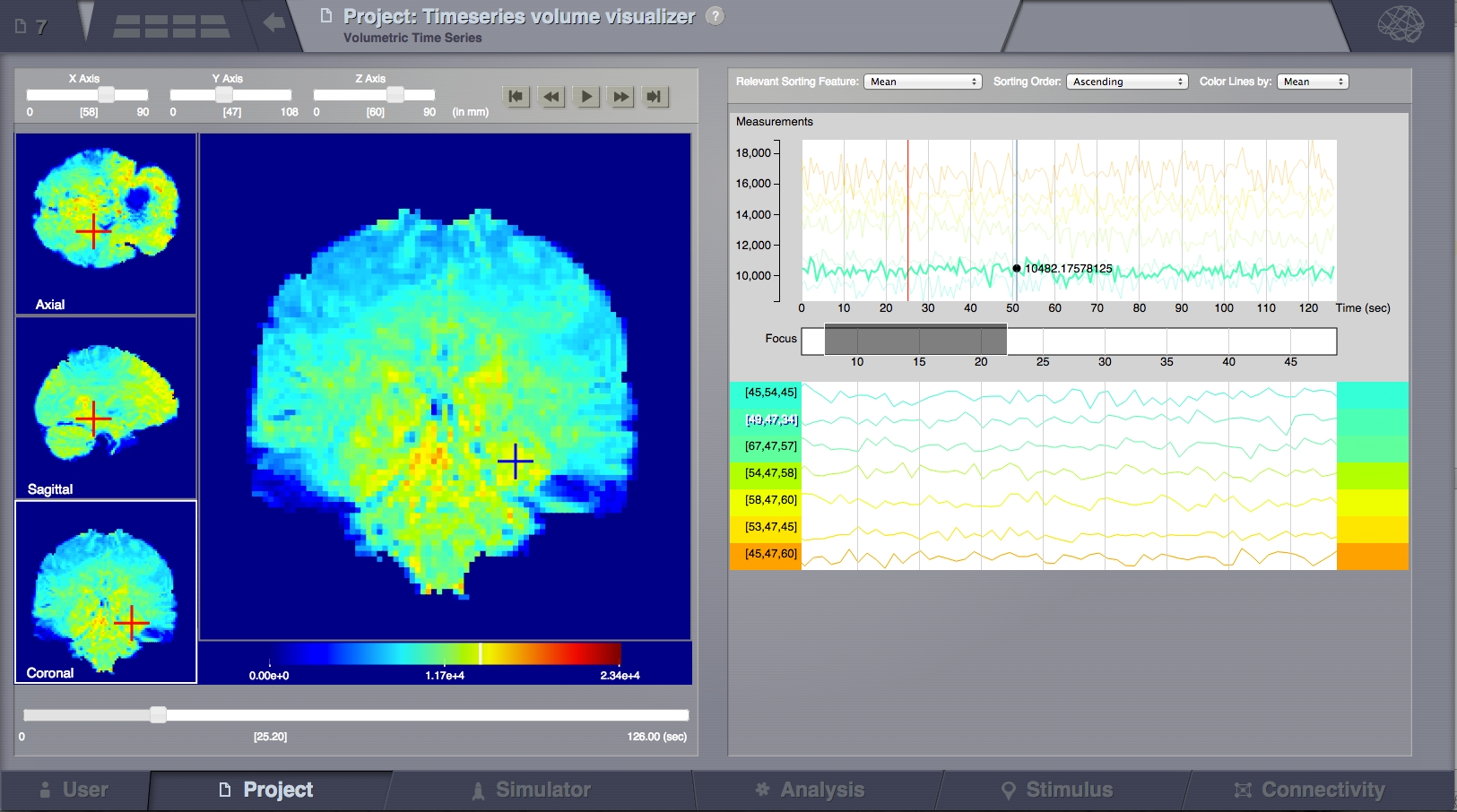
Time Series Volume with selections¶
Volumetric fragment
There are 3 navigation and viewing quadrants on the left and one main “focus quadrant” (left-central). It is possible to navigate in space using the slide controls on the top-left toolbar or by clicking on the 3 navigation quadrants on the most left part of the screen. So clicking in the 3 left squares will change the X, Y, Z of the planes slicing through the currently displayed volume (as the sliders on top are doing), while clicking in the main (central) square will select the clicked point for display of details on the right.
The crosses designate the selected voxel. It’s value is shown at the bottom of the focus quadrant. A white bar on the color legend also indicates this value.
The playback function is activated by clicking the play button on the top bar, and it will then change the display with time (left and right areas); The time series data is buffered from the server according to the currently section of view.
A different color map can be selected by clicked the Brain call-out in the top-right side of the screen. You might want to use the trim middle values feature with this viewer. It renders values around the mean transparent in the view. Also to be found on the Brain call-out.
Time Series Line Fragments
This is the right part of the TimeSeries Volume visualizer and is composed of other sub-parts:
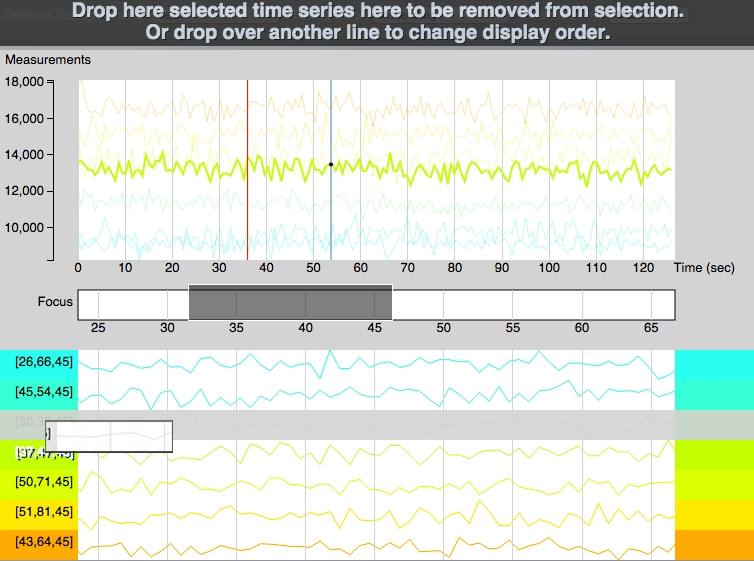
Time Series Volume Line Fragments¶
Global Time Series Graph
All selected lines are shown here (top area), with the same scaling. Some transparency is applied to the lines and only one line is highlighted at a time. Highlighting can be done by passing the mouse over the line on the global graph or by clicking the selected line in the sortable graphs bellow. Vertical scaling is done based only on the selected values and not on the complete data set. A red vertical line shows the current time point (correlated with the movie in TimeSeries Volume section). A blue line follows the mouse showing the value of the highlighted line at each point.
Time slice selection (focus):
This function can be used to display only a portion of the data, zooming on it bellow. Try dragging in this region. The grey selection box can be moved and resized. If the focused data looks flat, increase the selected window length. The selection will automatically set itself around the current time point with a default extent during playback.
Sortable Graphs:
Every selected time series from the volume is shown on a separate line and labeled based on its coordinates from the 3D space. Adding lines in this section can be done by clicking in the left area on the main quadrant.
Display multi dimensional time series:
In case the Time Series displayed comes from a TVB simulation, when the Neuronal Mass Model supports multiple modes and state-variables, then it is necessary to choose what to display, as this viewer can only show 2D results. To choose from Mode and State Variable dimensions, a selector will appear on the top-right area. When changing the selection, the coloring for the left-side volume regions will change accordingly.
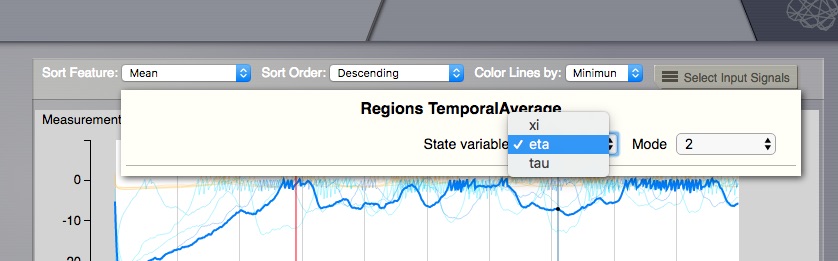
Time Series Volume - select when multiple dimensions¶
Already selected Time series lines on the right, will remain unchanged, when Mode and State Variable change, but if you click again on the left side volume, new lines will be added, for the currently active Mode and State Variable. One can inspect in the line title, the details for that point (including X, Y, Z position in the volume, full region name, Mode and State Variable).
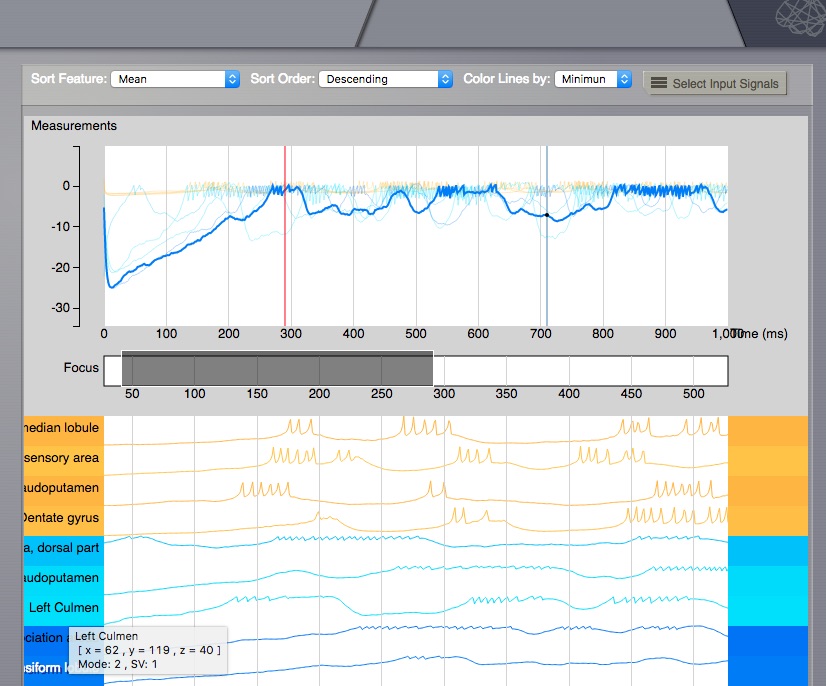
Time Series Volume - Line title¶
Important notice:
While these time lines share the temporal axis they do not share the vertical one. The signal amplitudes are dynamically scaled so as to make the signal features visible. Amplitudes are not comparable among two of these signals.
The lines are colored following the selected feature in “Color Lines by” at the top of the screen. They are then sorted automatically by one of the selected methods or manually, by dragging and dropping each line in the desired position, as seen on the picture bellow. Lines can be removed by dragging them to the top “trash bin area” that appears every time a line is selected to be dragged.
Connectivity Measure Visualizer¶
This visualizer can be used for displaying various Brain Connectivity Measures, related to a given Connectivity.
On the X axis, we will see the Connectivity nodes listed, and for each of them, we see the computed measure on the Y axis.
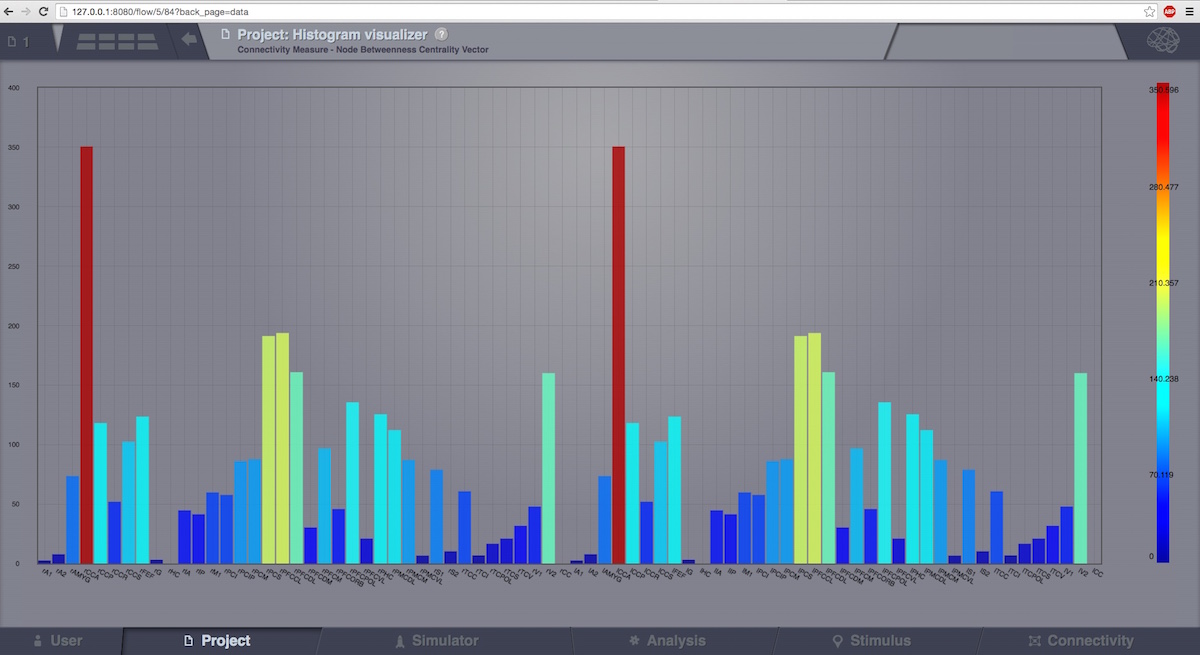
Connectivity Measure Visualizer.¶
Topographic Visualizer¶
This visualizer can be used for displaying various Brain Connectivity Measures, related to a given Connectivity. Its input is the same as for the previous visualizer (Connectivity Measure Visualizer), but the display is completely different. Instead of a discrete view, this time, we can have a continous display (with gradients).
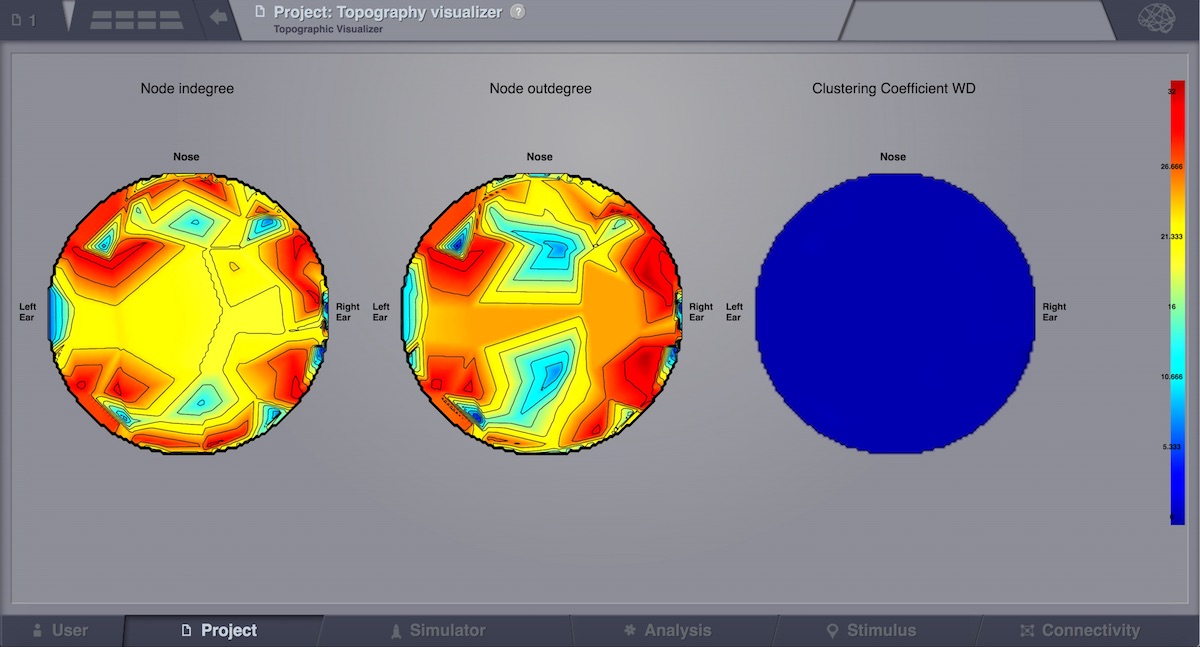
Preview for Topographic Visualizer¶
Surface Visualizer¶
This visualizer can be used for displaying various Brain Surfaces. It is a static view, mainly for visual inspecting imported surfaces in TVB. Optionally it can display associated RegionMapping entities for a given surface. Navigate the 3D scene like in the Brain Activity Visualizer.
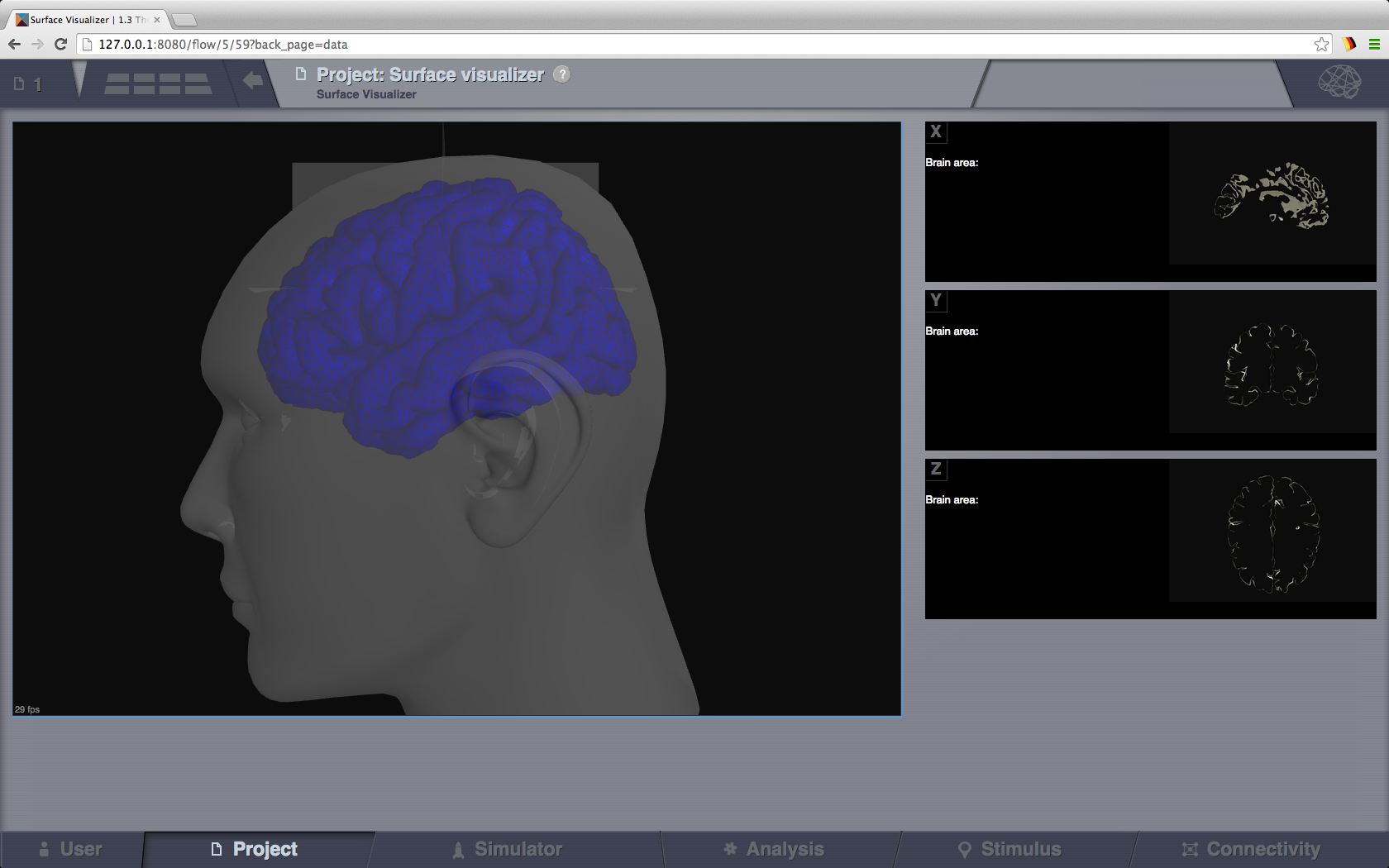
Surface Visualizer.¶
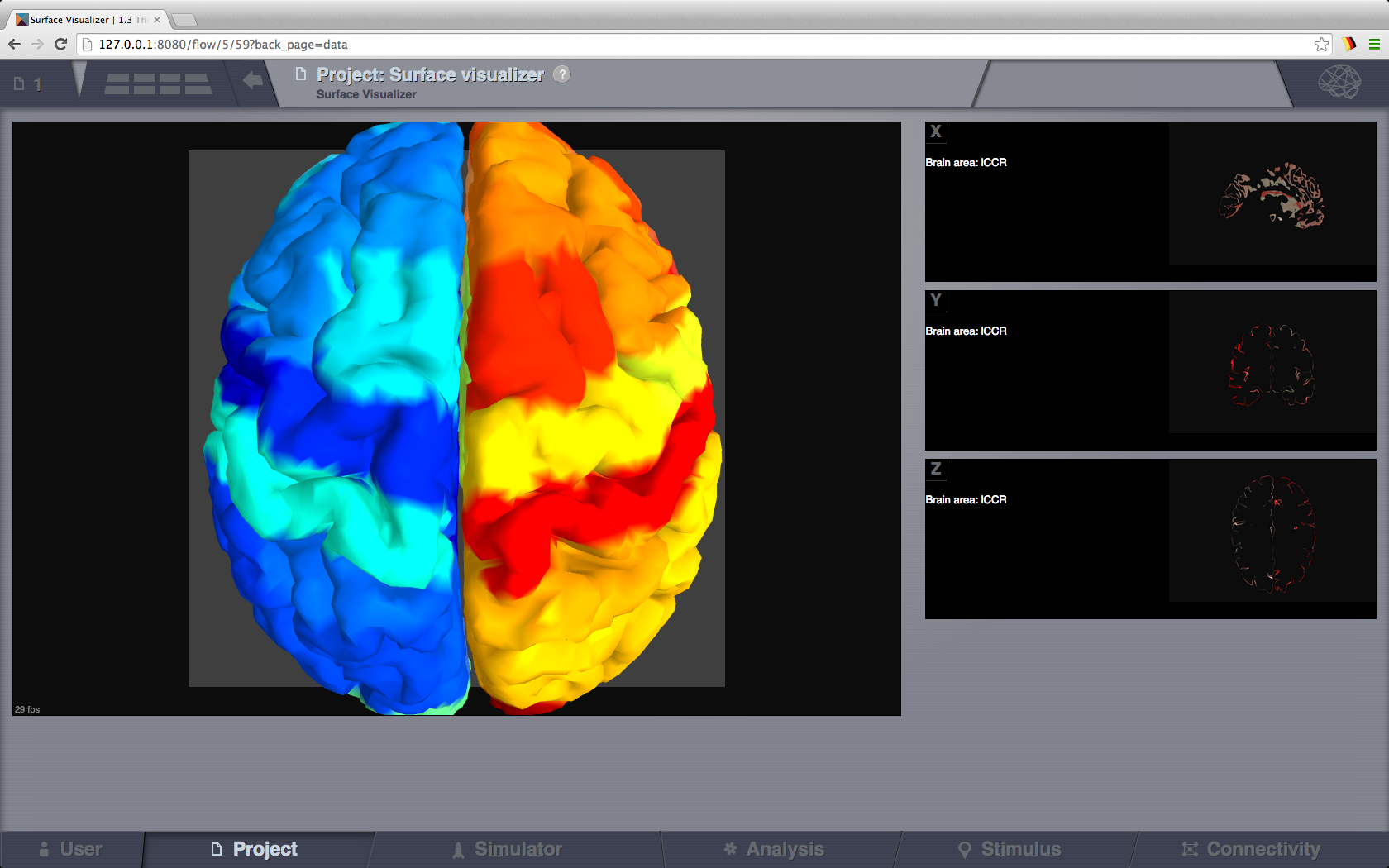
Cortical Surface Visualizer with Region Mapping applied.¶
Sensor Visualizer¶
This visualizer can be used for displaying EEG, MEEG, and internal sensors . It is a static view, intended for visual inspecting imported sensors in TVB. Optionally it can display the sensors on a EEG cap surface.
To show sensors displaying on a Cap, check the call-out on the top-right corner.
When displaying the EEG sensors on a EEG Cap surface, we are automatically computing a “parcellation”. Currently this parcellation has no anatomical meaning, it is only based on distance (a vertex gets coloured as the closest sensor).
Navigate the 3D scene like in the Brain Activity Visualizer.
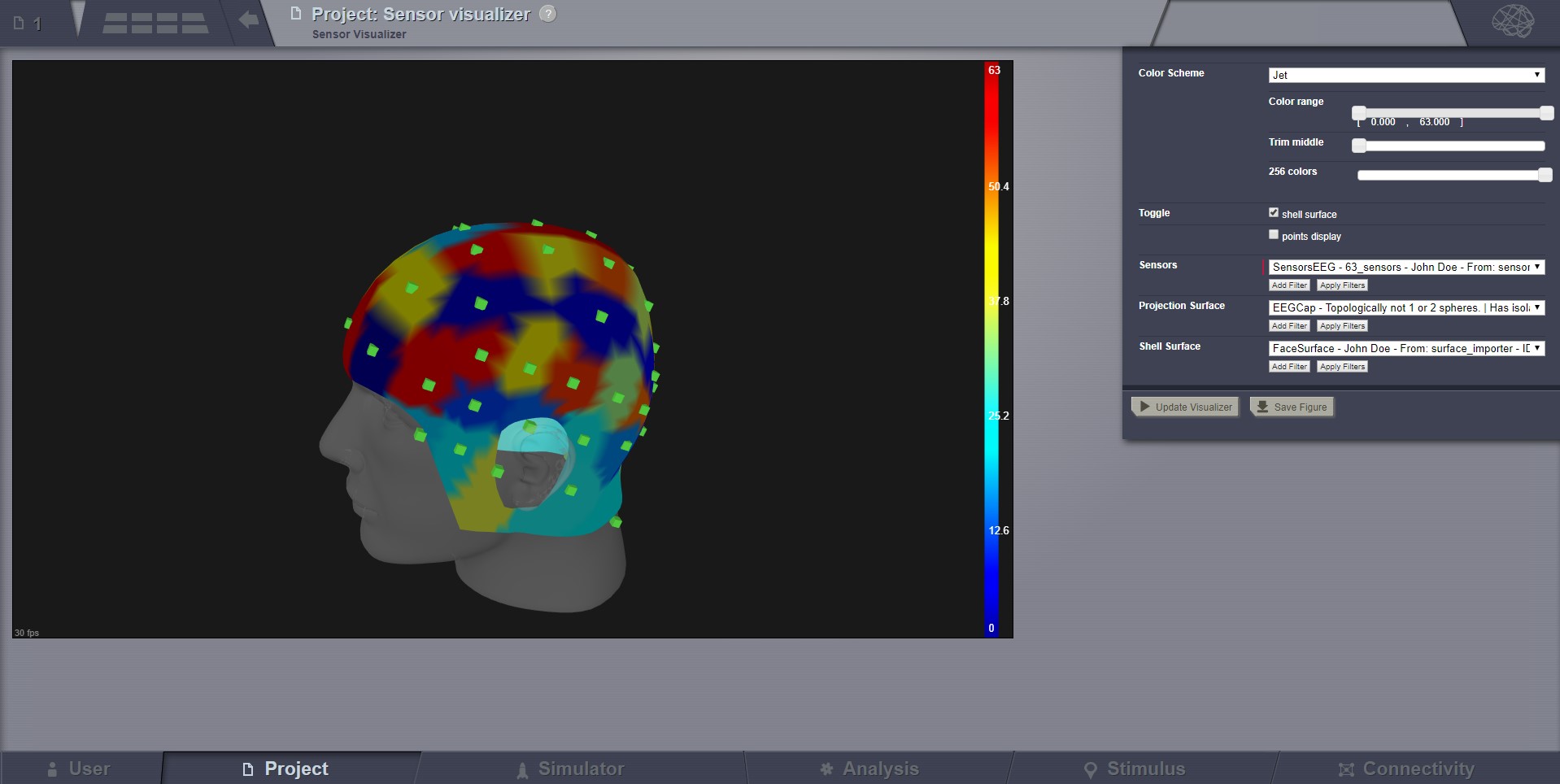
EEG Sensors.¶
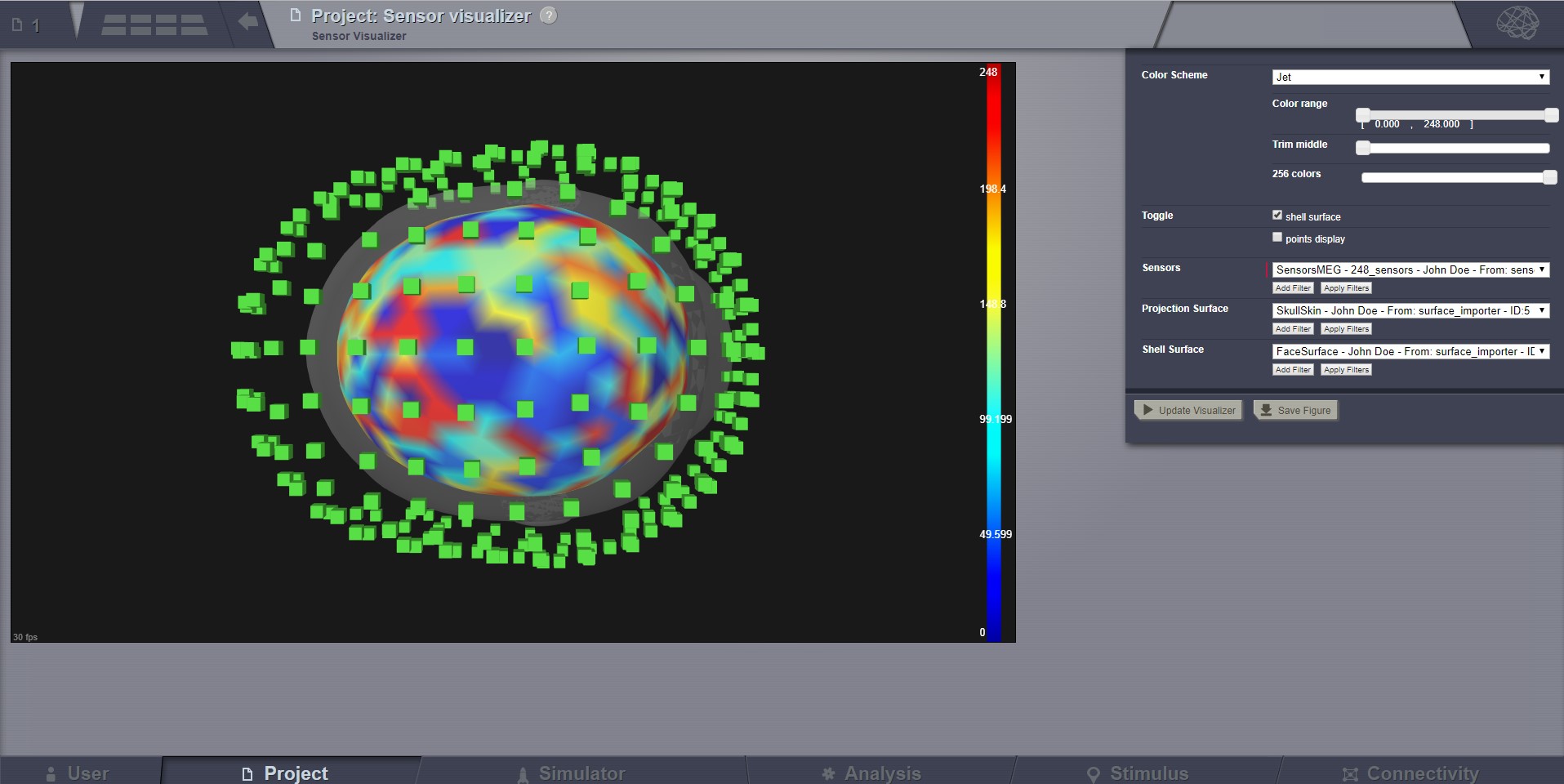
MEG Sensors.¶
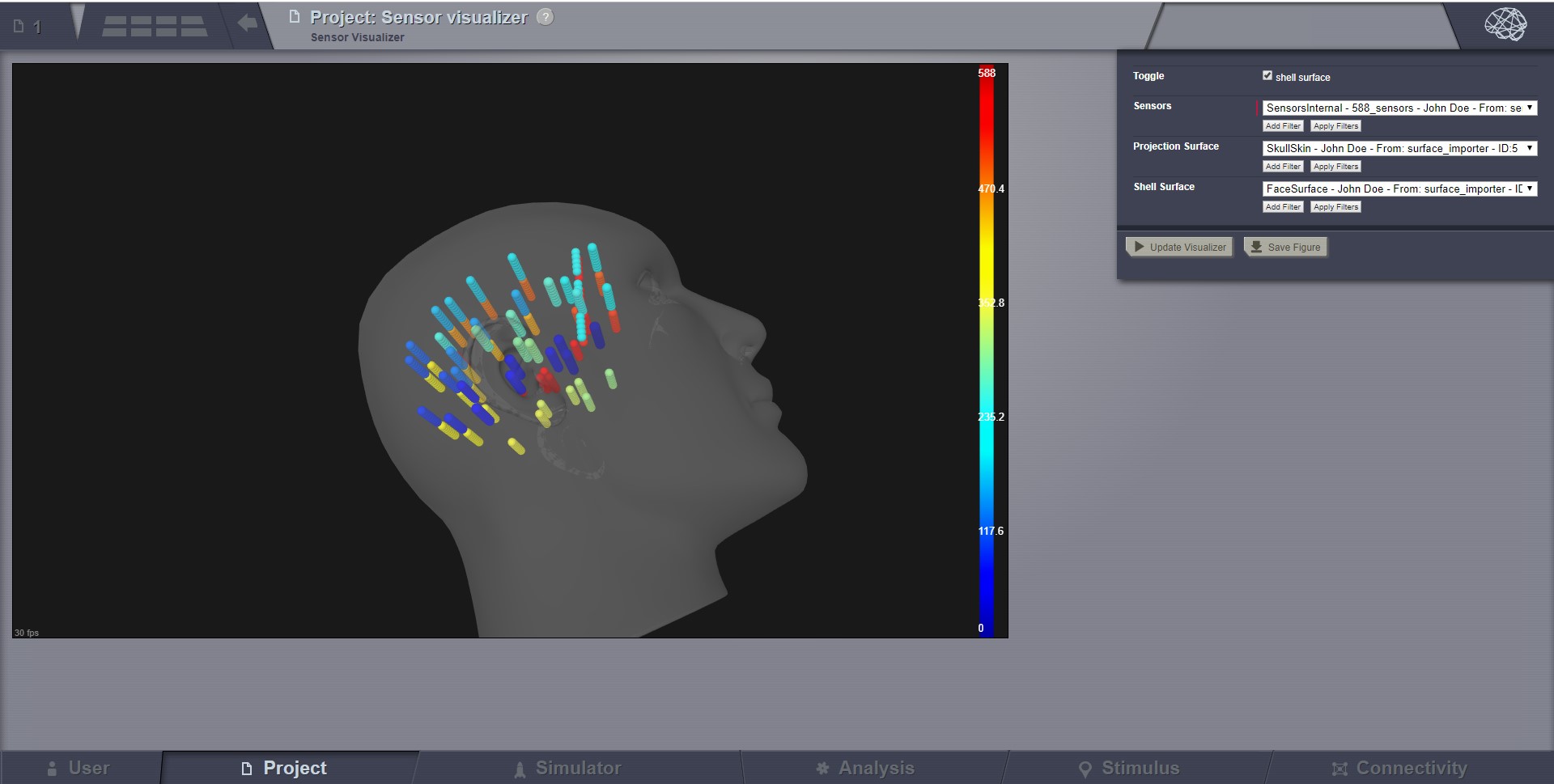
Internal Sensors.¶
Local Connectivity Visualizer¶
Once a Local Connectivity DataTypes (which in fact is a huge sparse matrix of max size surface vertices x surface vertices, shaped after the cut-off) gets computed, one can view the correlation of a given vertex compared to all its neighbours, by launching this viewer. The Local Connectivity viewer can be launched from the DataType overlay (after clicking on a Local Connectivity datatype, and then selecting TAB Visualizers), or from Connectivity (bottom page menu), Local Connectivity option on top of the page, then select an existing LocalConnectivity and finally click “View” from the right side menu.
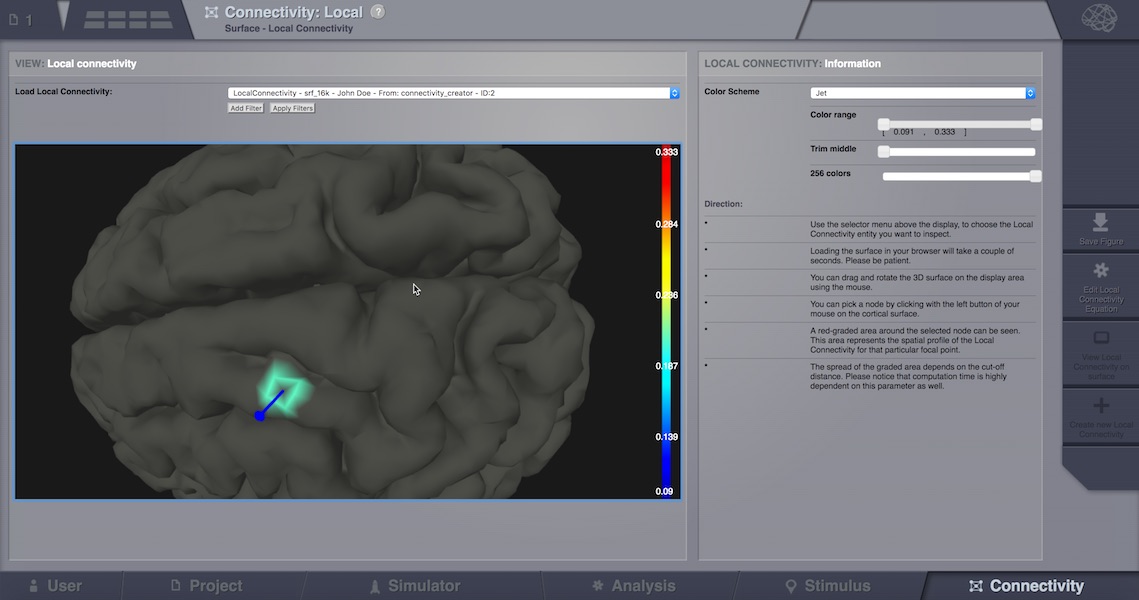
Inspect local connectivity on surface.¶
In order to see actual correlations, one should pick (by mouse click) a vertex on the 3D cortical surface once it loads in the canvas. The colors displayed nearby, show connected vertices with the selected point.
Annotations Visualizer¶
- This viewer shows ontology annotations linked with TVB connectivity regions. It is composed of two main display areas:
3D left-side canvas with TVB regions. These regions are color coded, based on the connectivity region index (similar to Surface Visualizer when a Region Mapping entity is selected). From the most top-right corner menu, you can change the color scheme used to draw these regions coloring.
2D tree display of ontology annotations. A tooltip will appear if you go with the mouse over various nodes, and will show you details imported from the ontology.
- The two areas (left and right) are linked, both ways:
You can pick a vertex in 3D and have the corresponding tree node highlighted on the right-side, or backwards:
Click on the tree, and have the corresponding region(s) highlighted in 3D.
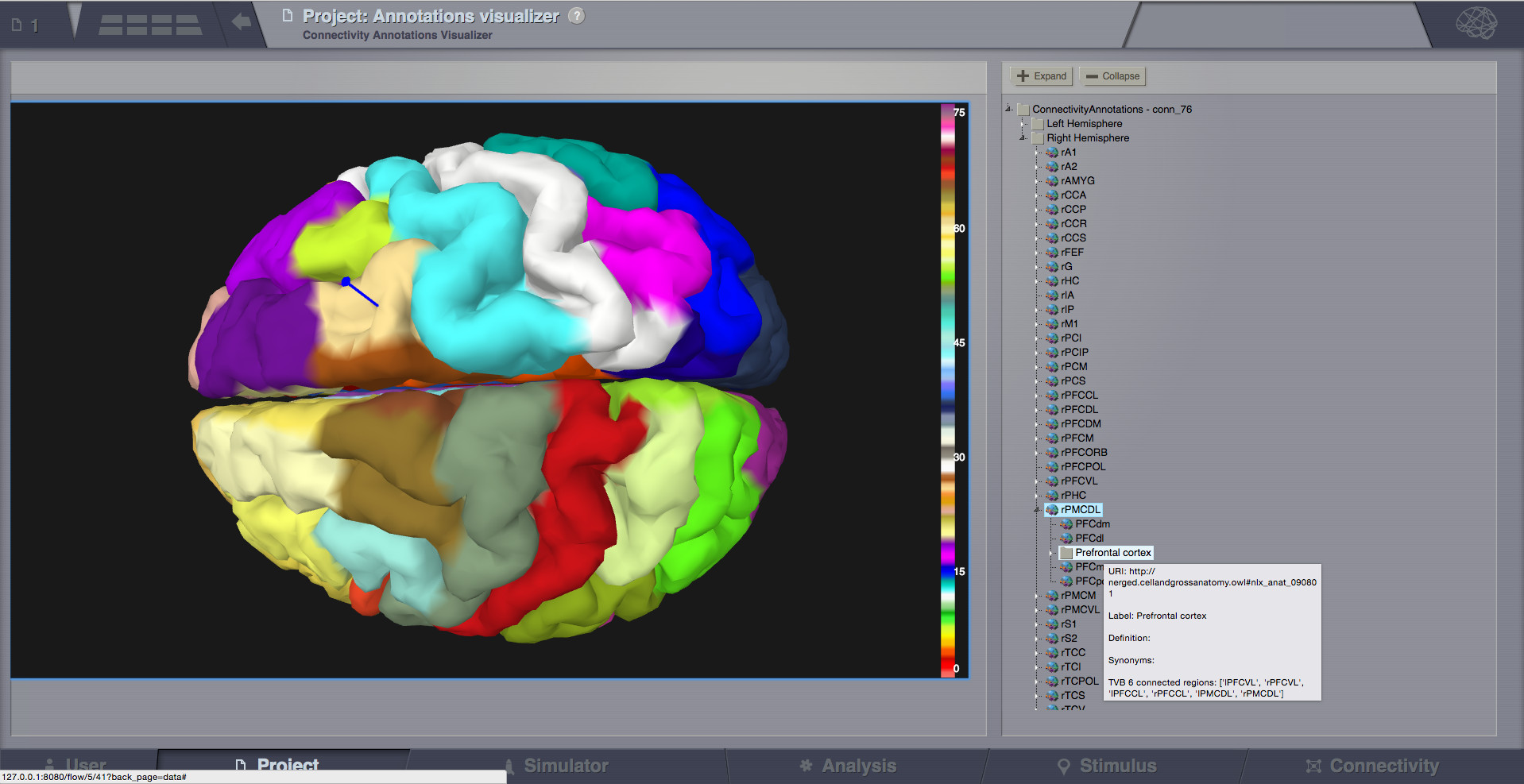
Pick a vertex in 3D and have the corresponding tree node selected on the right.¶
- Hints:
There is a checkbox on the top-right menu to draw region boundaries in the 3D canvas
When you click on an ontology node on the right, a message text will appear on the top area of the page, telling you how many TVB regions are linked to this ontology term
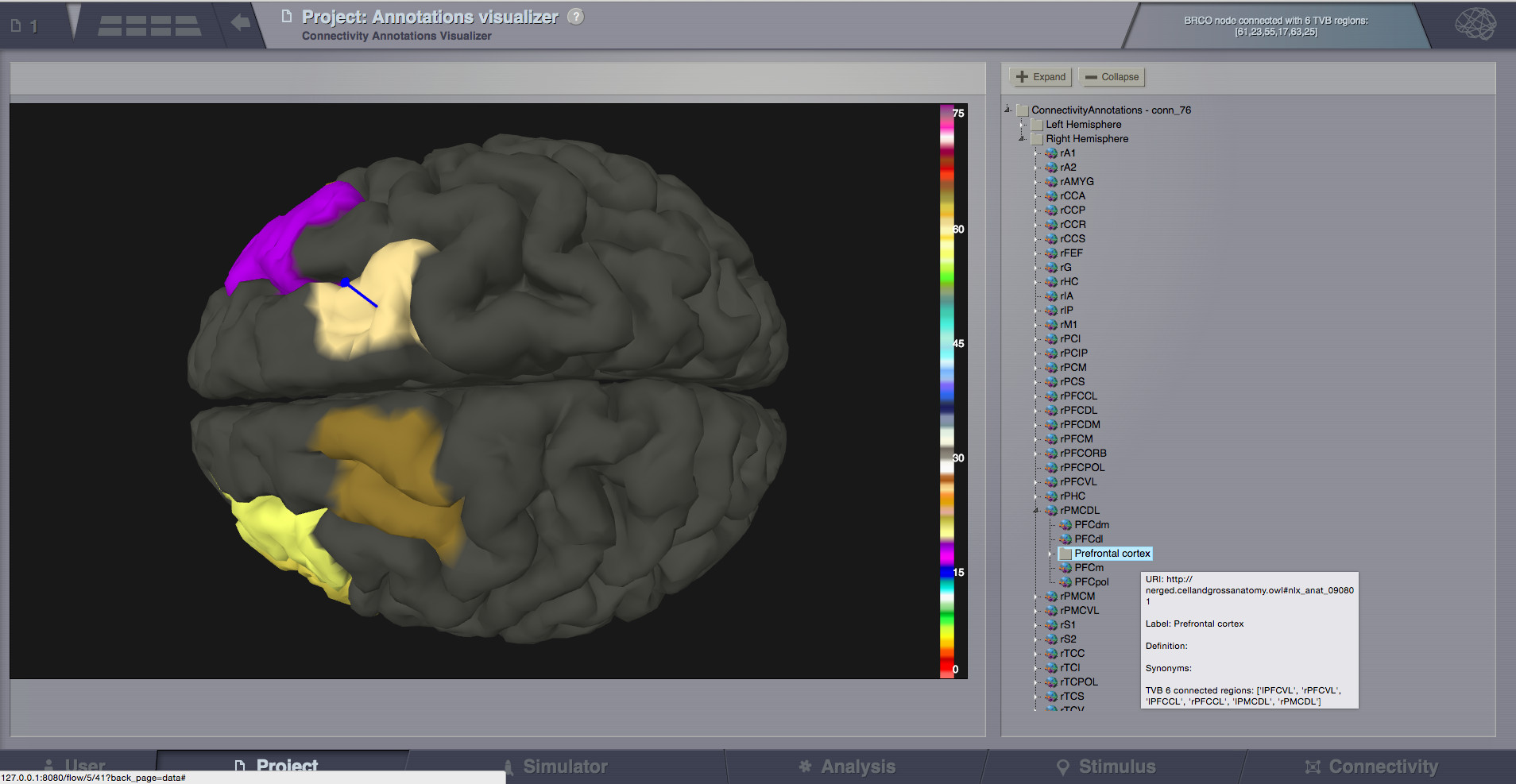
Select a tree node on the right, and have the linked regions highlighted in 3D.¶
Group Display¶
Discrete PSE Visualizer¶
Discrete Parameter Space Exploration (PSE) View will show up information on multiple simulation results at once.
In TVB it is possible to launch multiple simulations by varying up to 2 input parameters (displayed on the X and Y axis of the current viewer). Each simulation result has afterwards “metrics” computed on the total output. Each metric is a single number. Two metrics are emphasized in this viewer in the node shapes and node colors.
When moving with your mouse cursor over a graph node, you will see a few details about that particular simulation result. When clicking a node, an overlay window will open, which gives you full access to view or further analyze that particular Simulation result.
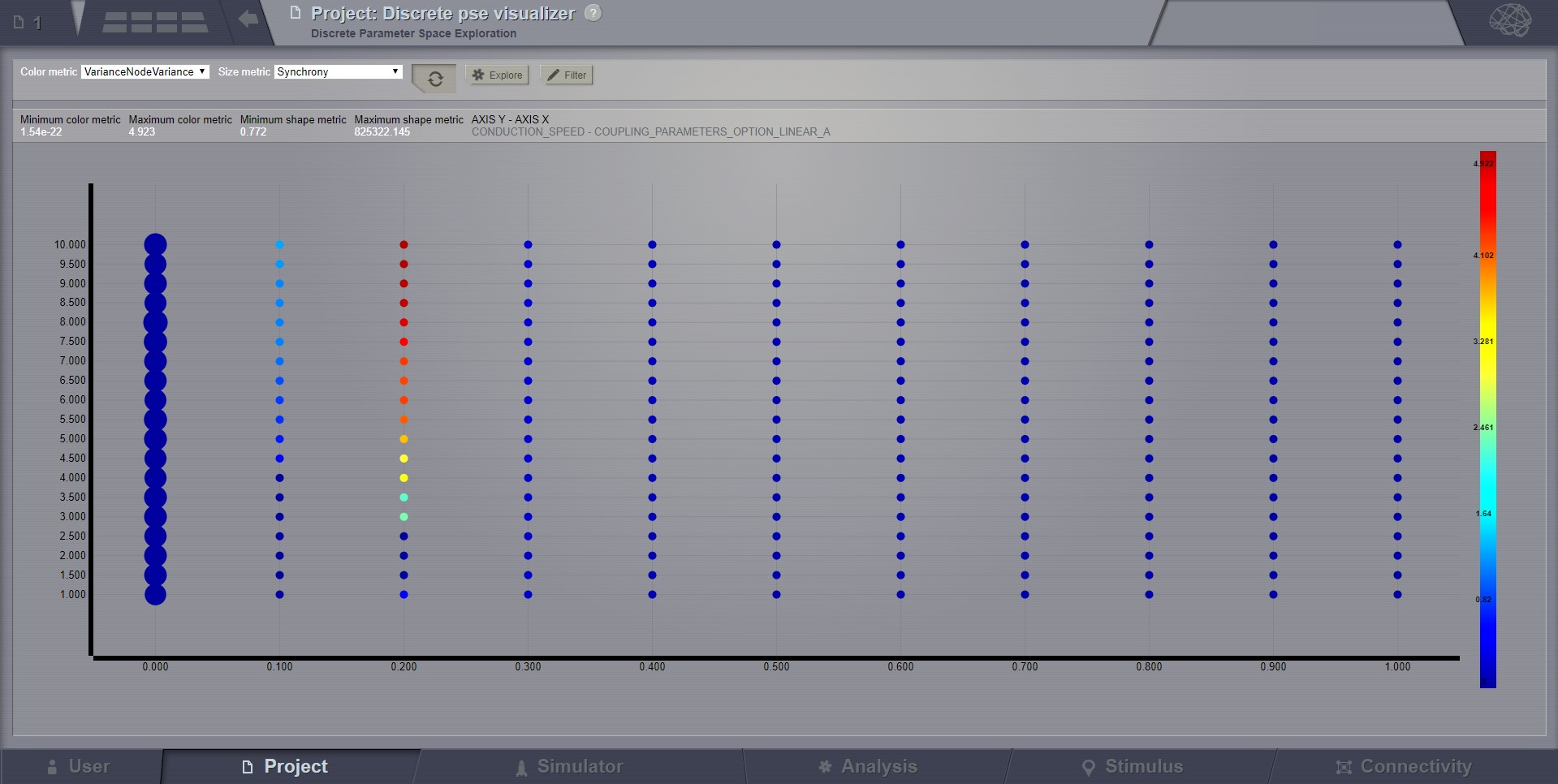
Preview for Discrete PSE Visualizer, when varying two input parameters of the simulator¶
A newly incorporated feature is the option to pan the canvas in/out or left/right/up/down. To pan you may click and drag on top of one of the axes, and to zoom in double click or out shift + double click. This will allow the inspection of very large batch simulations section by section. The same mouse over, and clicking rules apply from above.
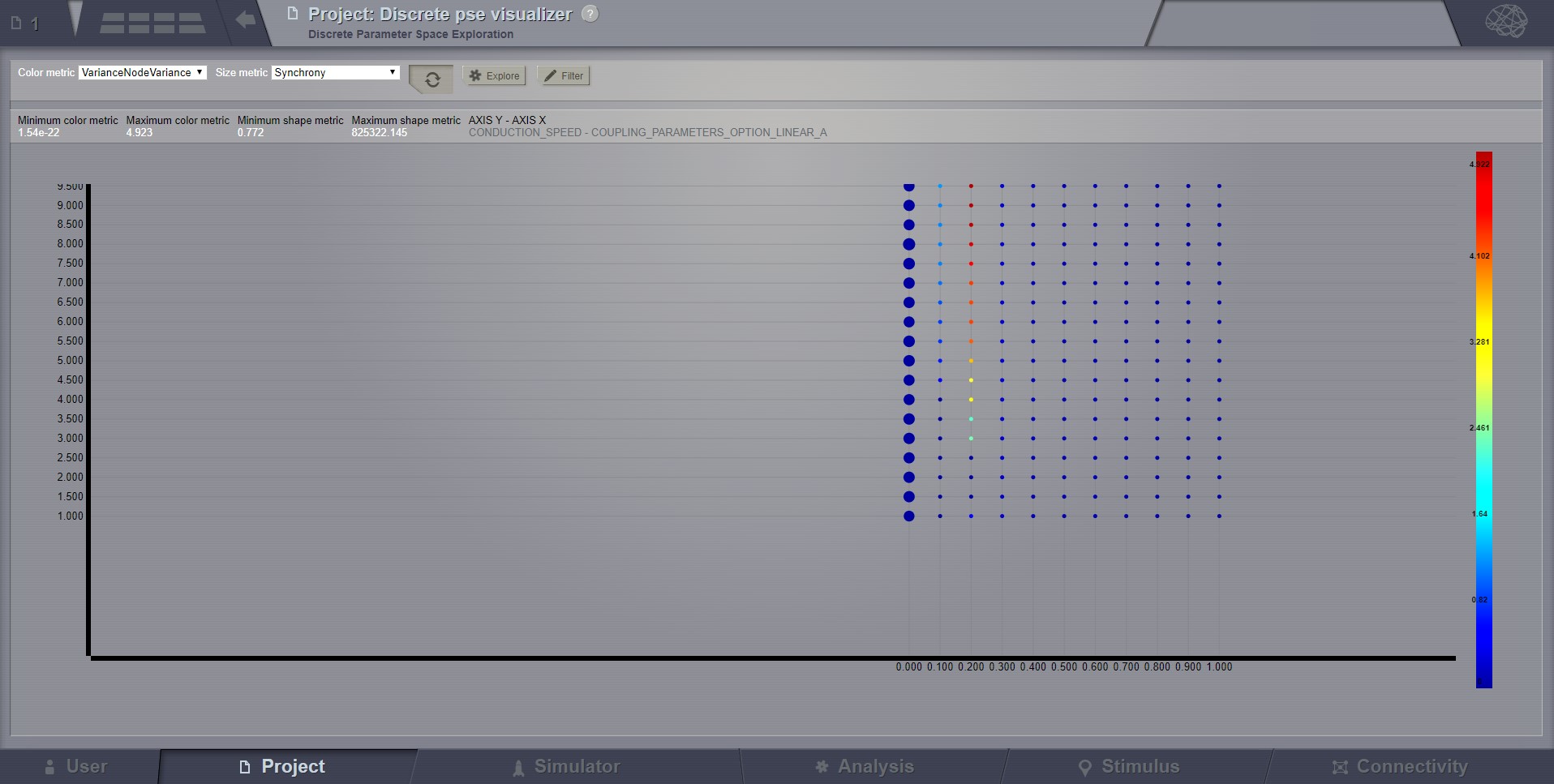
Panning the Graph¶
The next new tool is the filter button. This allows users to specify threshold values for either the color or size metric and render results transparent if they are below that value. This tool has the option to invert the threshold rule which makes the results above that threshold transparent instead. Also, the user has the choice to make their filter more specific by adding further criteria rows that relate to the one which came before it through selected logical operators (AND OR). It is worth noting that in order to perform filtering that requires grouping of the logical operations ([foo and bar] or baz) as different from (foo and [bar or baz]) sequential filters must be applied: one filter execution then the other.
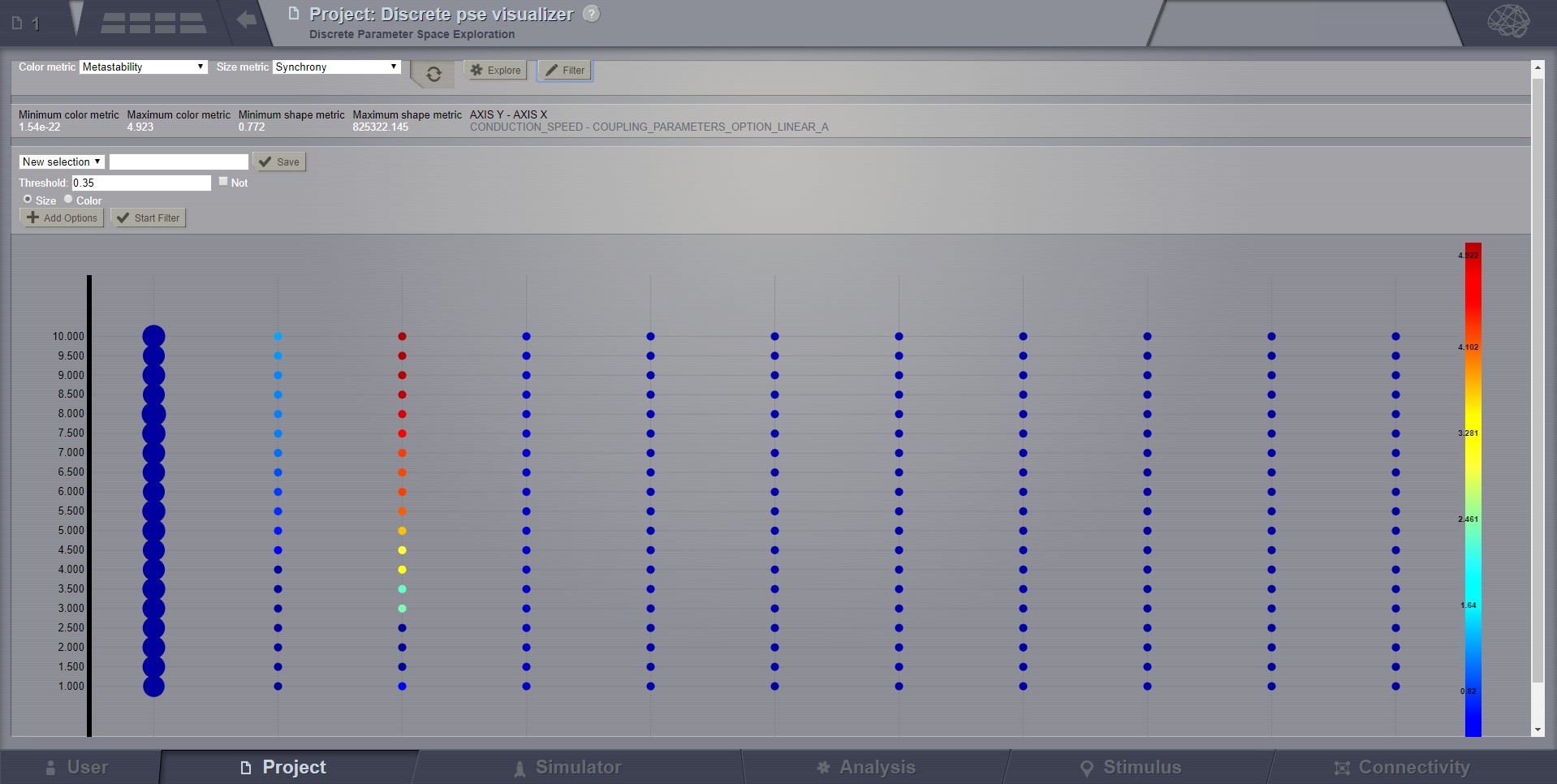
Filter support¶
(The Explore button is currently disabled because this functionality is not fully implemented)
The last tool to be described in the PSE Discrete Viewer is the Explore tool. This tool is meant to give users the option to select regions of the Parameter Space to be filled in with new results. Currently only the front end of this tool is complete, so upon clicking the explore button the mouse cursor becomes a cross hair, and sections of the graph can be selected. Upon creation of this selection, grid lines are placed to demonstrate where new results would be added given the user’s chosen step values. To adjust these values simply drag the sliders in the drop down menu for the explore tool, and the grid lines will adjust until they suit the user.
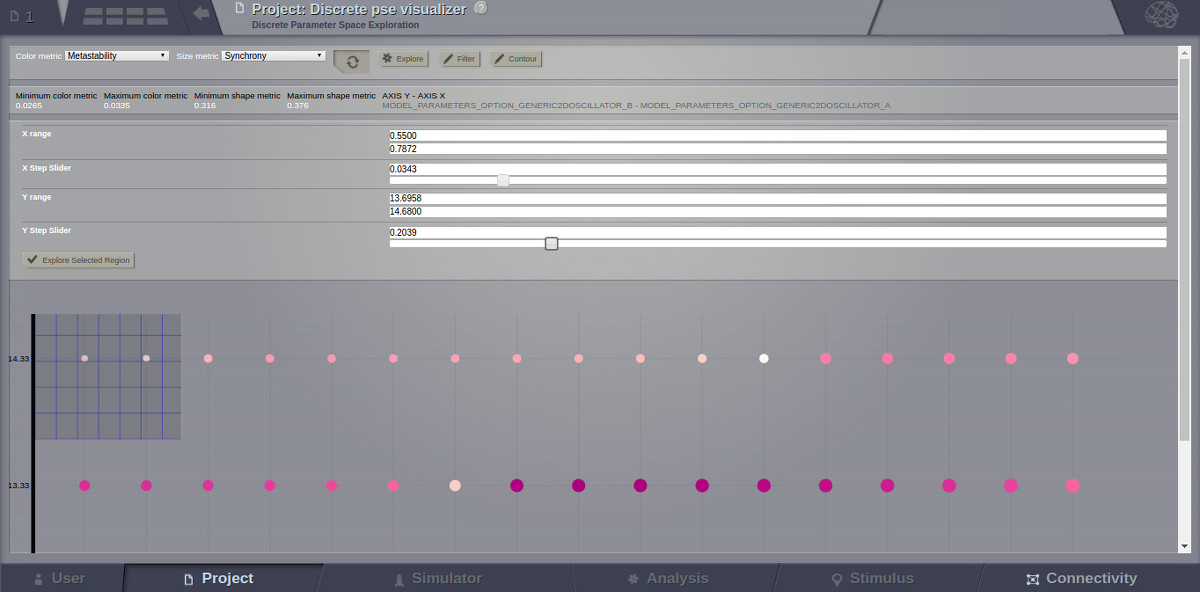
Explore Tool, with Region Selected¶
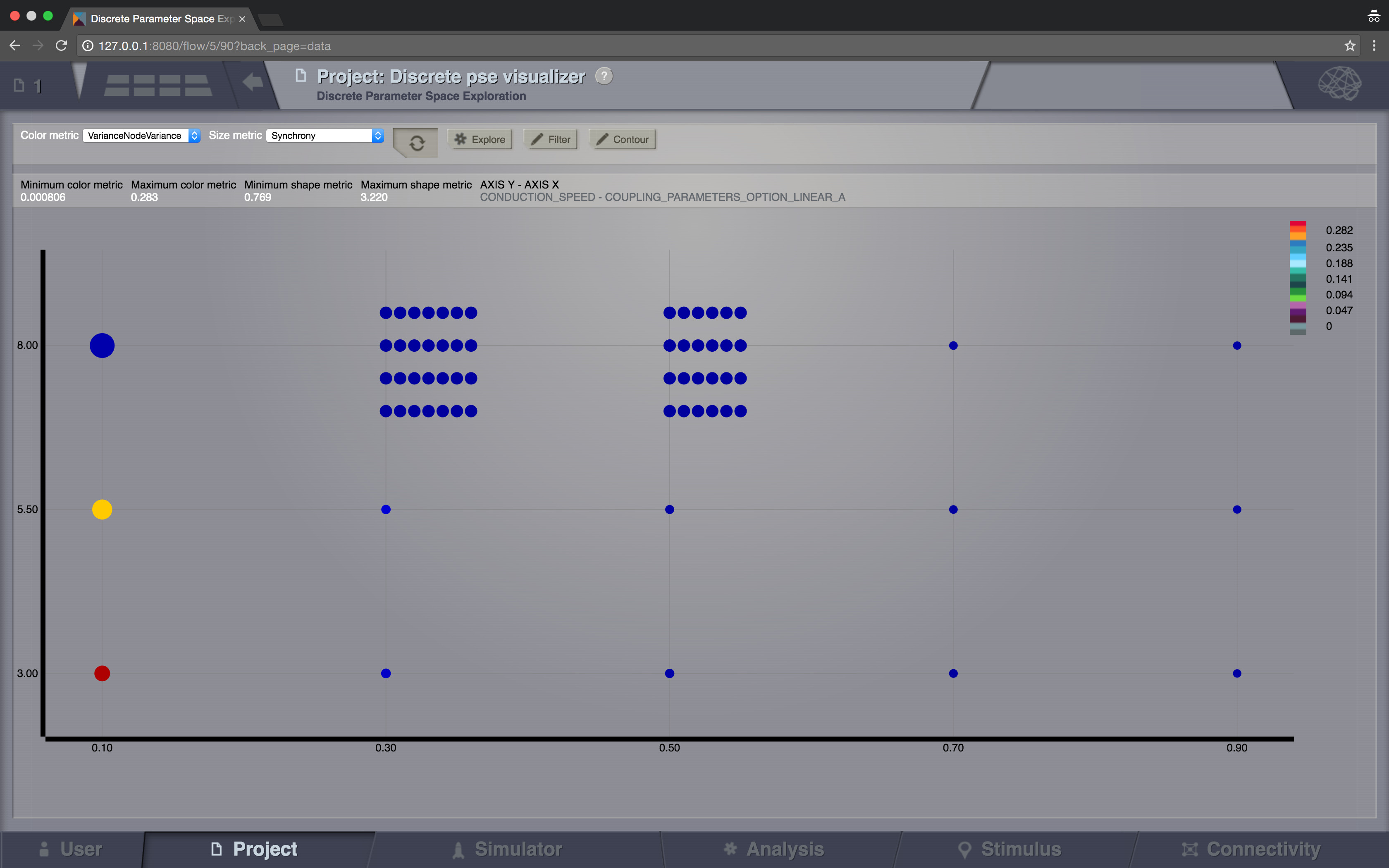
Sparse results¶
Isocline PSE Visualizer¶
Continuous Parameter Space Exploration View, will show the effect of varying Simulator parameters in a continuous form.
When running a range of Simulations in TVB, it is possible to do it by varying up to 2 input parameters (displayed on the X and Y axis of current viewer). This visualizer supports ranges with 2 dimensions only, it does not support ranges with only one dimension. Also both varying dimensions need to be numeric parameters (no DataType ranges are supported for display in this visualizer).
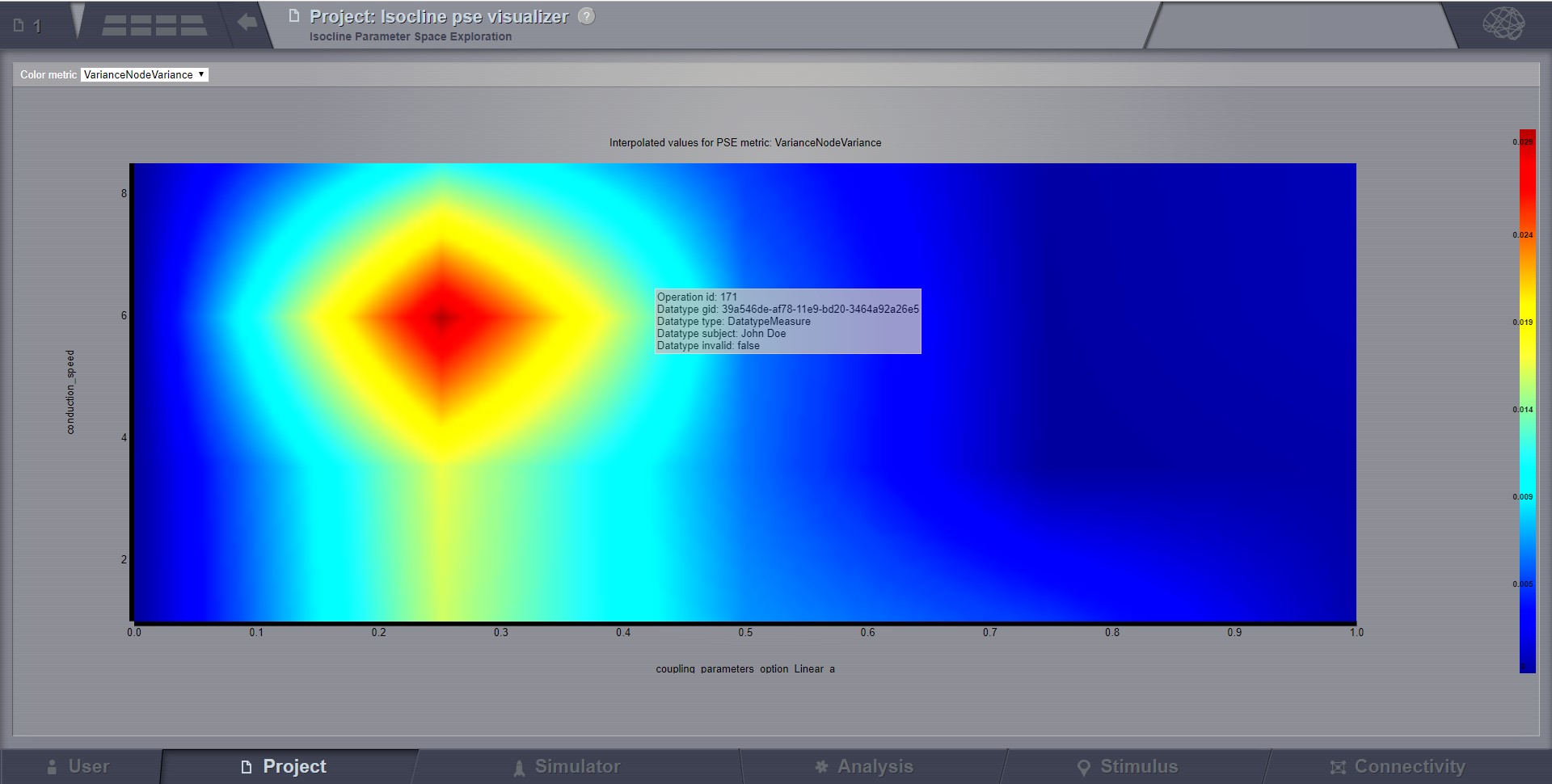
Preview for Continuous PSE Visualizer, when varying two numeric input parameters of the simulator¶
Controls for scaling or zooming the graph are available in this viewer. When you click on the coloured area, an overlay window will open, having the possibility to view or further analyze the simulation result closest to the point where you clicked.
Analyzers + Visualizers¶
Covariance Visualizer¶
Displays the covariance matrix. The matrix size is number of nodes x number of nodes
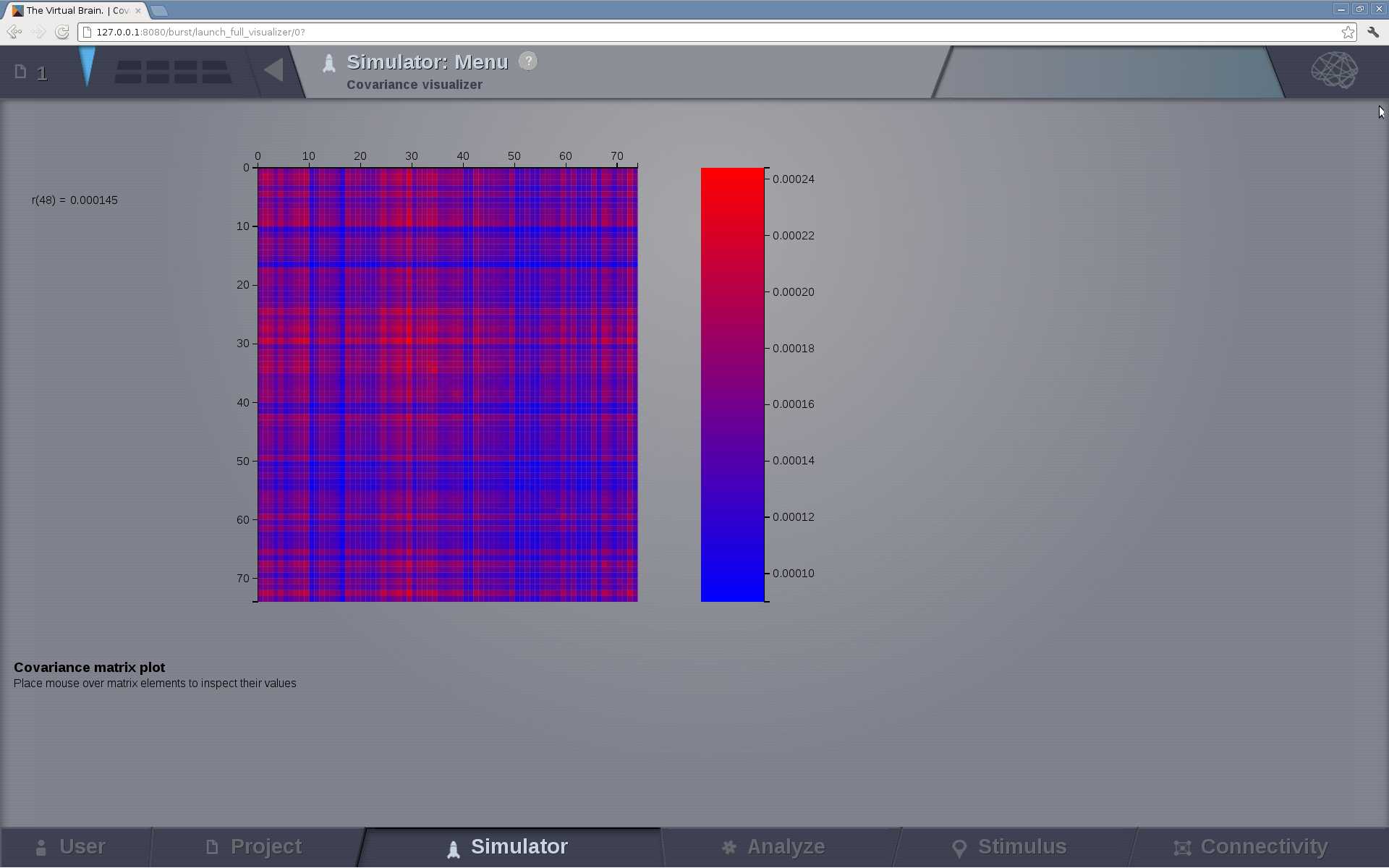
Preview for Covariance Visualizer¶
Cross Coherence Visualizer¶
Displays the cross-coherence matrix. Axes represent brain nodes. The matrix size is number of nodes x number of nodes.
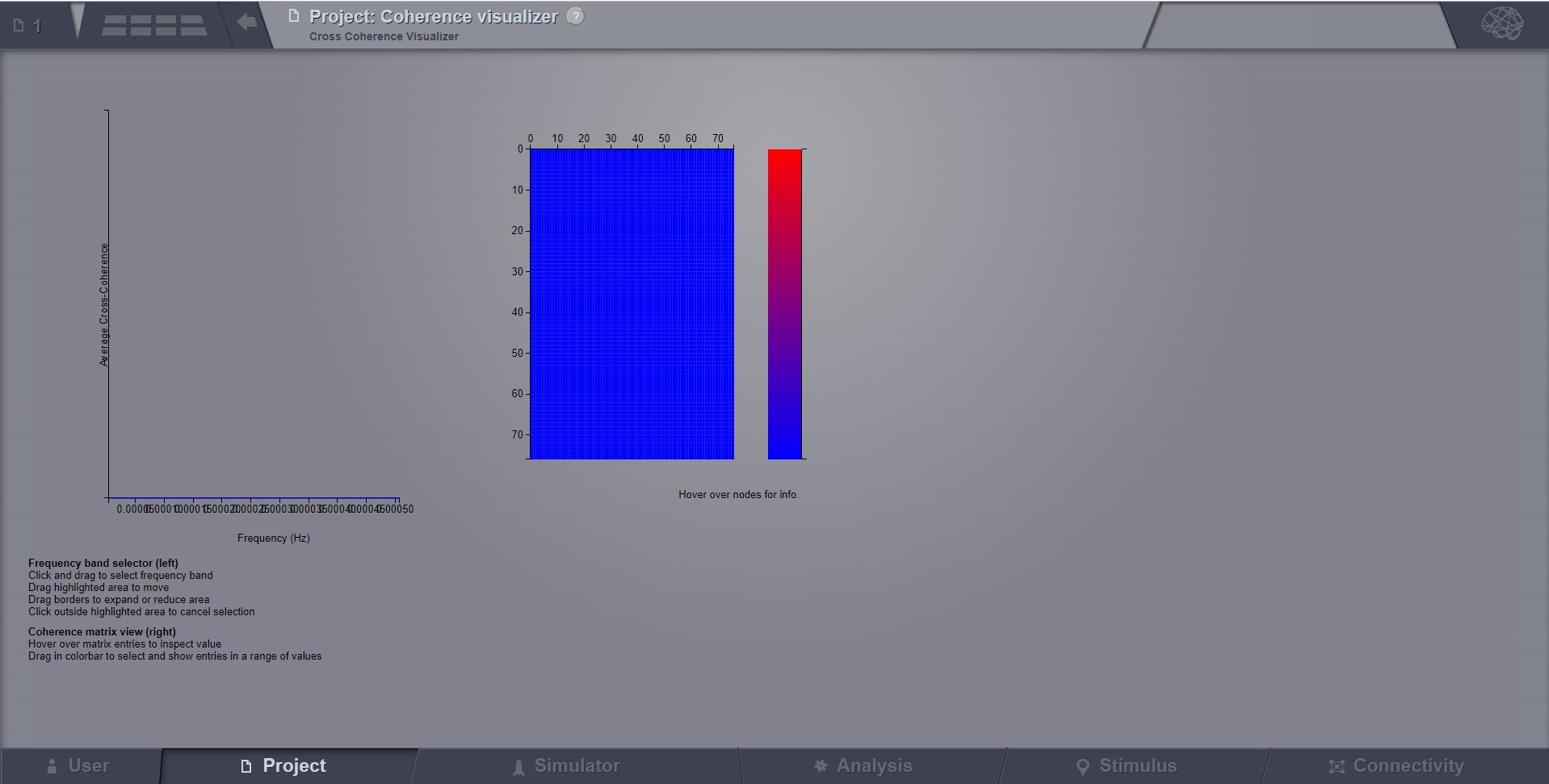
Preview for Cross Coherence Visualizer¶
Complex Coherence Visualizer¶
Displays the complex-cross-coherence matrix. Axes represent brain nodes. The matrix is a complex ndarray that contains the number of nodes x number of nodes cross spectrum for every frequency and for every segment. The thick line represents the Mean and the colored area the SD of CohSpec.
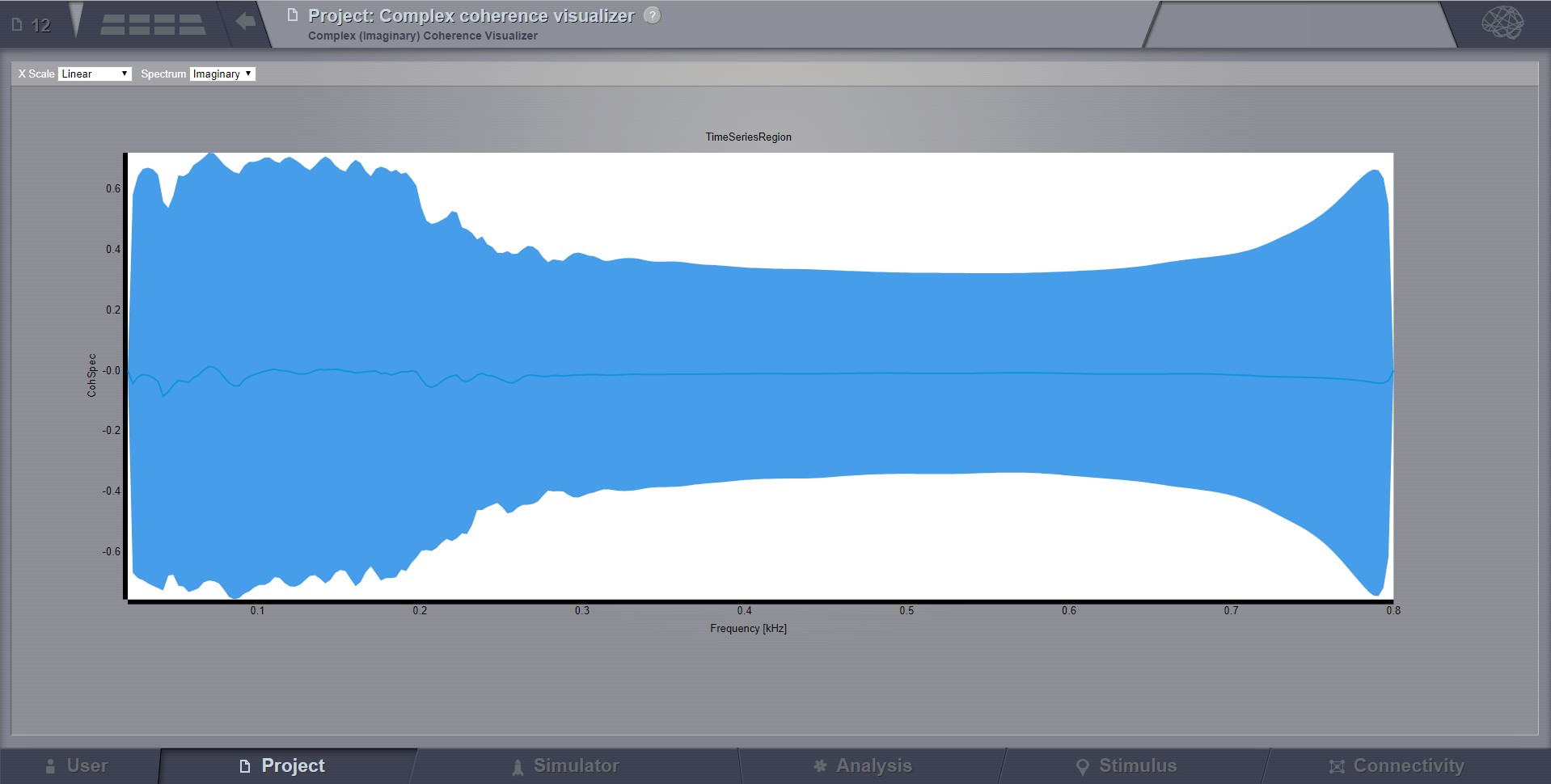
Preview for Complex Coherence Visualizer¶
Cross Correlation Visualizer¶
Displays the cross-correlation matrix. It is similar to the previous matrix visualizers.
Pearson Coefficients Visualizer¶
Displays the Pearson cross correlation coefficients matrix. As the correlation matrix is symmetric, only half of it is actually displayed.
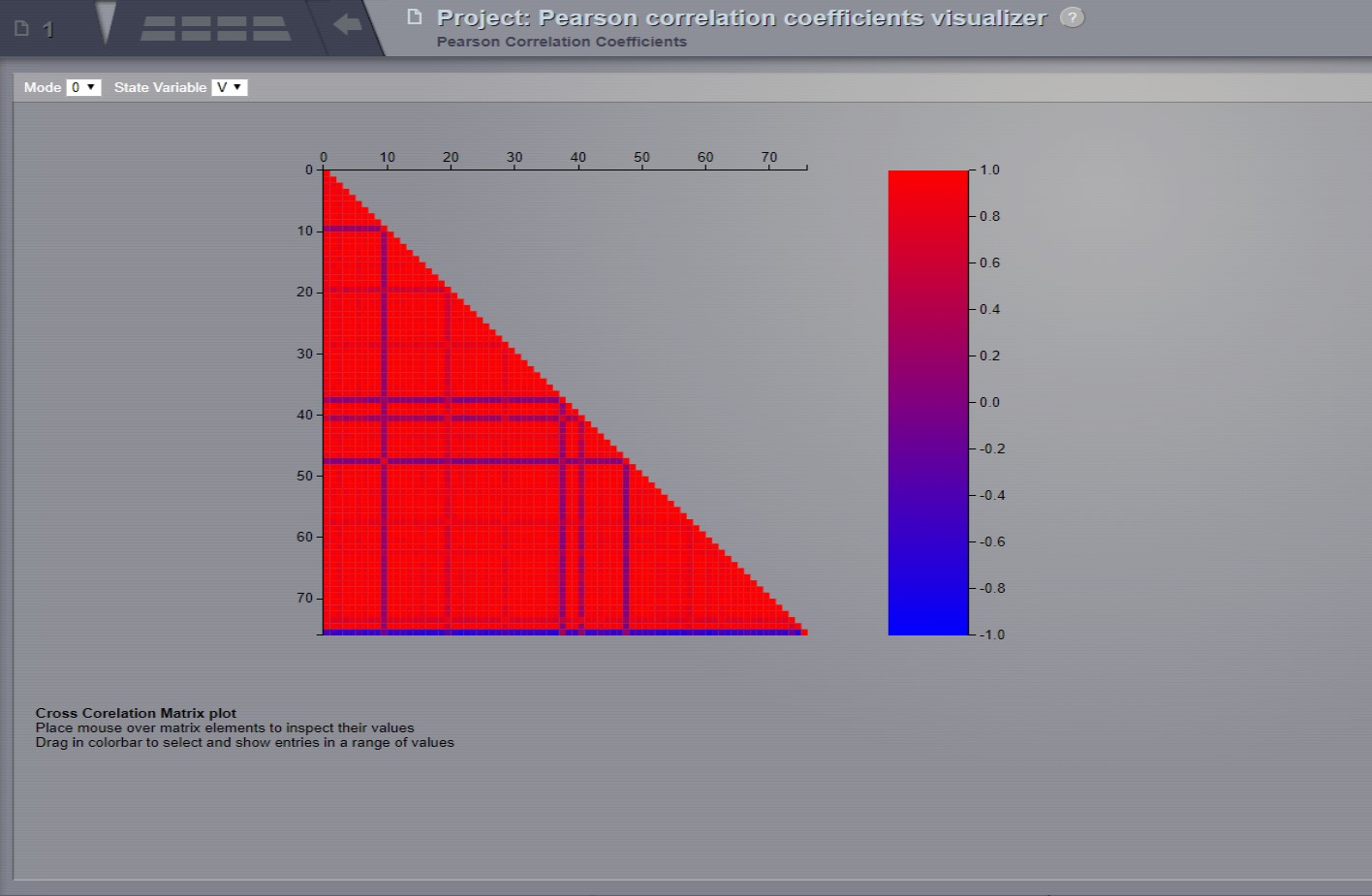
Preview for Pearson Cross Correlation Visualizer¶
Fourier Spectrum Visualizer¶
Plots the power spectrum of all nodes in a time-series.
From the top bar, you can choose the scale (logarithmic or linear) and when the resulted Timeseries has multiple modes and State variables, choose which one to display. After you change a selection in this top bar, the viewer will automatically refresh.
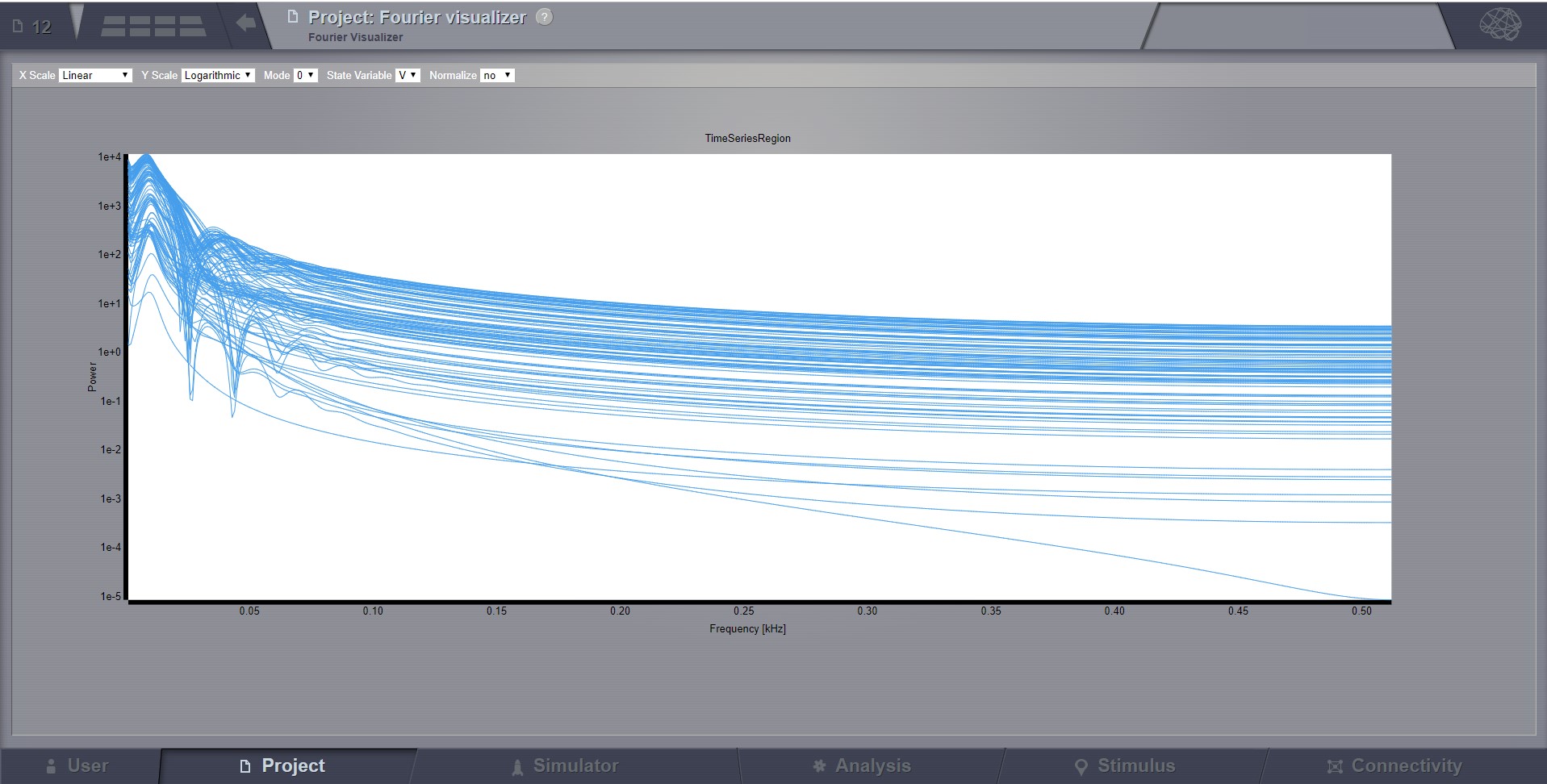
Preview for Fourier Spectrum Visualizer¶
Principal Component Visualizer¶
On the left, the ring plot displays the fraction of the variance that is explained by each component.
On the right, the first ten components are plotted against the brain nodes (variables).
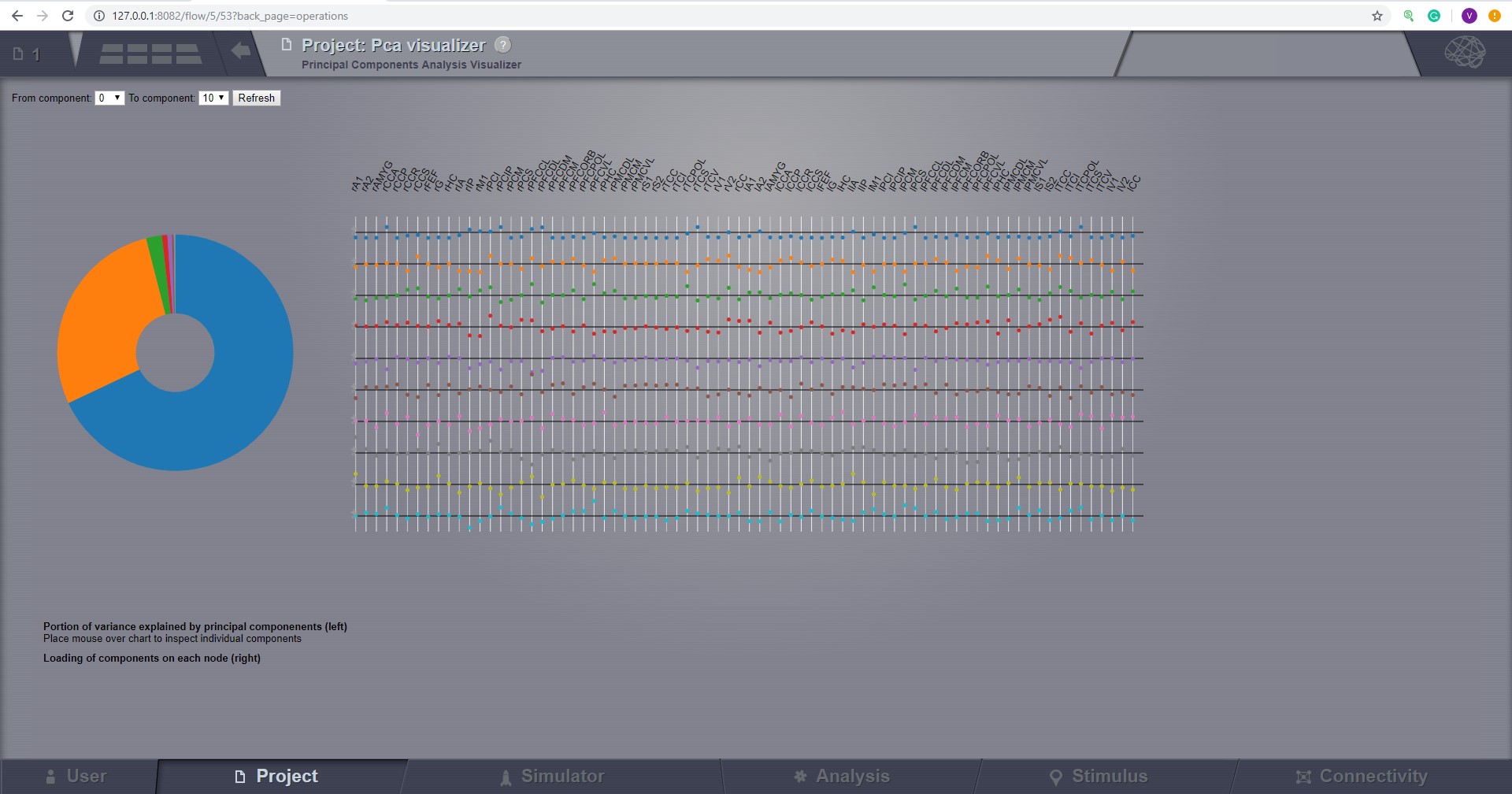
Preview for Principal Components Analysis Visualizer¶
Independent Component Visualizer¶
ICA takes time-points as observations and nodes as variables.
As for PCA the TimeSeries datatype must be longer (more time-points) than the number of nodes. Mostly a problem for TimeSeriesSurface datatypes, which, if sampled at 1024Hz, would need to be greater than 16 seconds long.
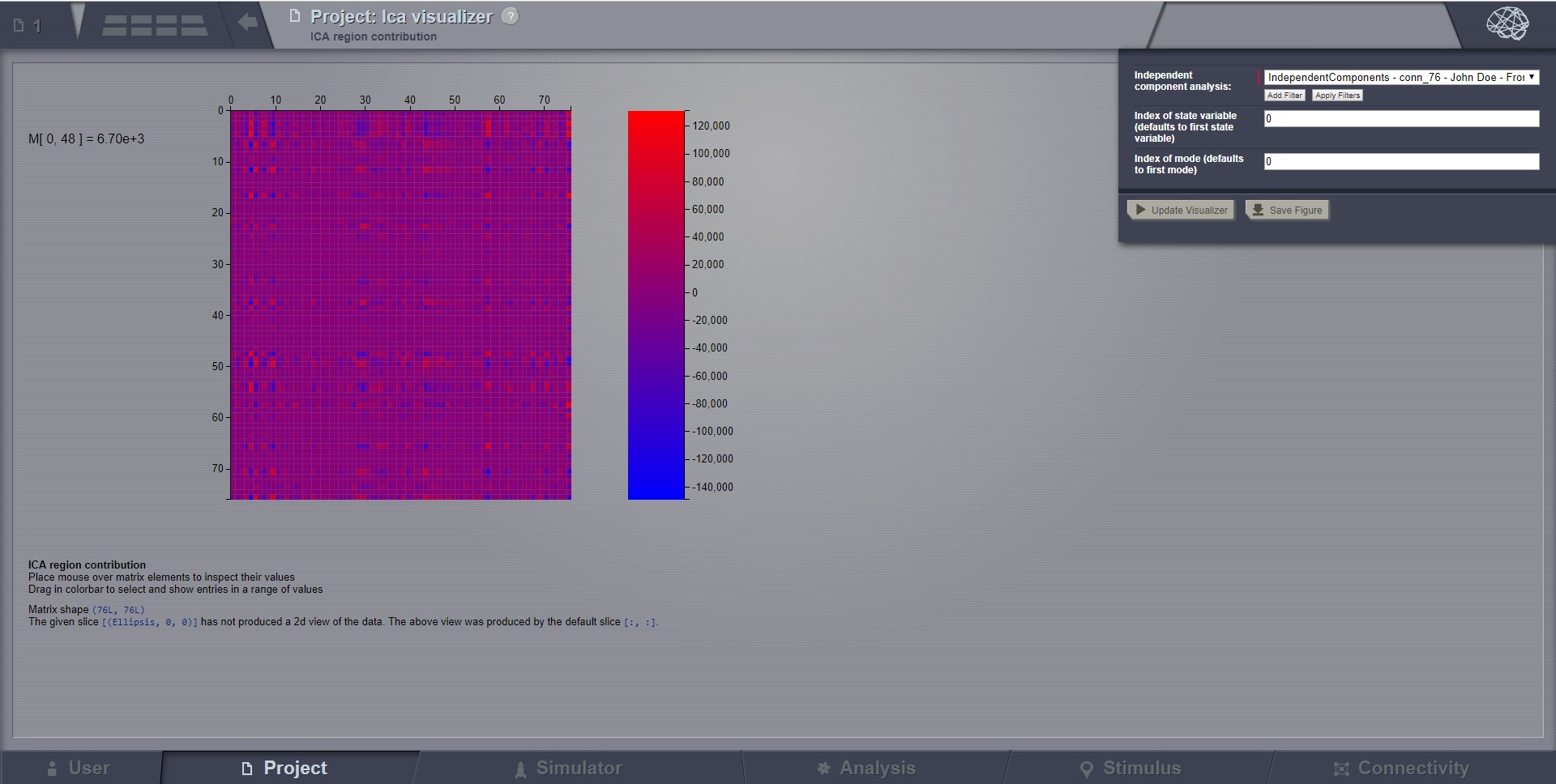
Preview for Independent Components Analysis Visualizer¶
Wavelet Spectrogram Visualizer¶
2D representation that shows how the signals wavelet spectral coefficients (frequency) vary with time.
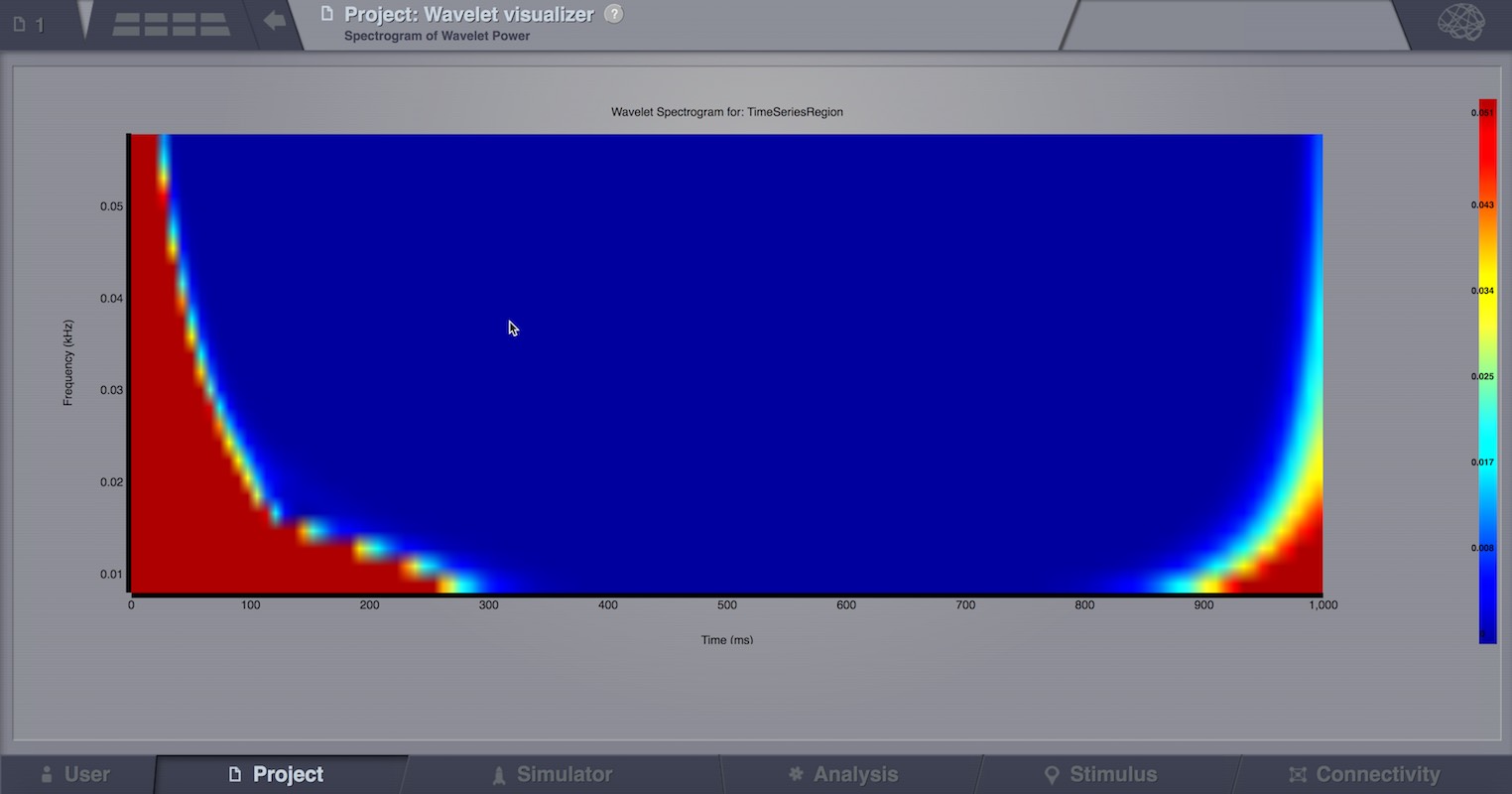
Preview for Wavelet Visualizer¶
Matrix Visualizer¶
This is a 2D representation of a generic matrix-like result.
In case the current Datatype has more than 2 dimensions, the ND array will be cut, with a default slice. The user can also input the slice manually.
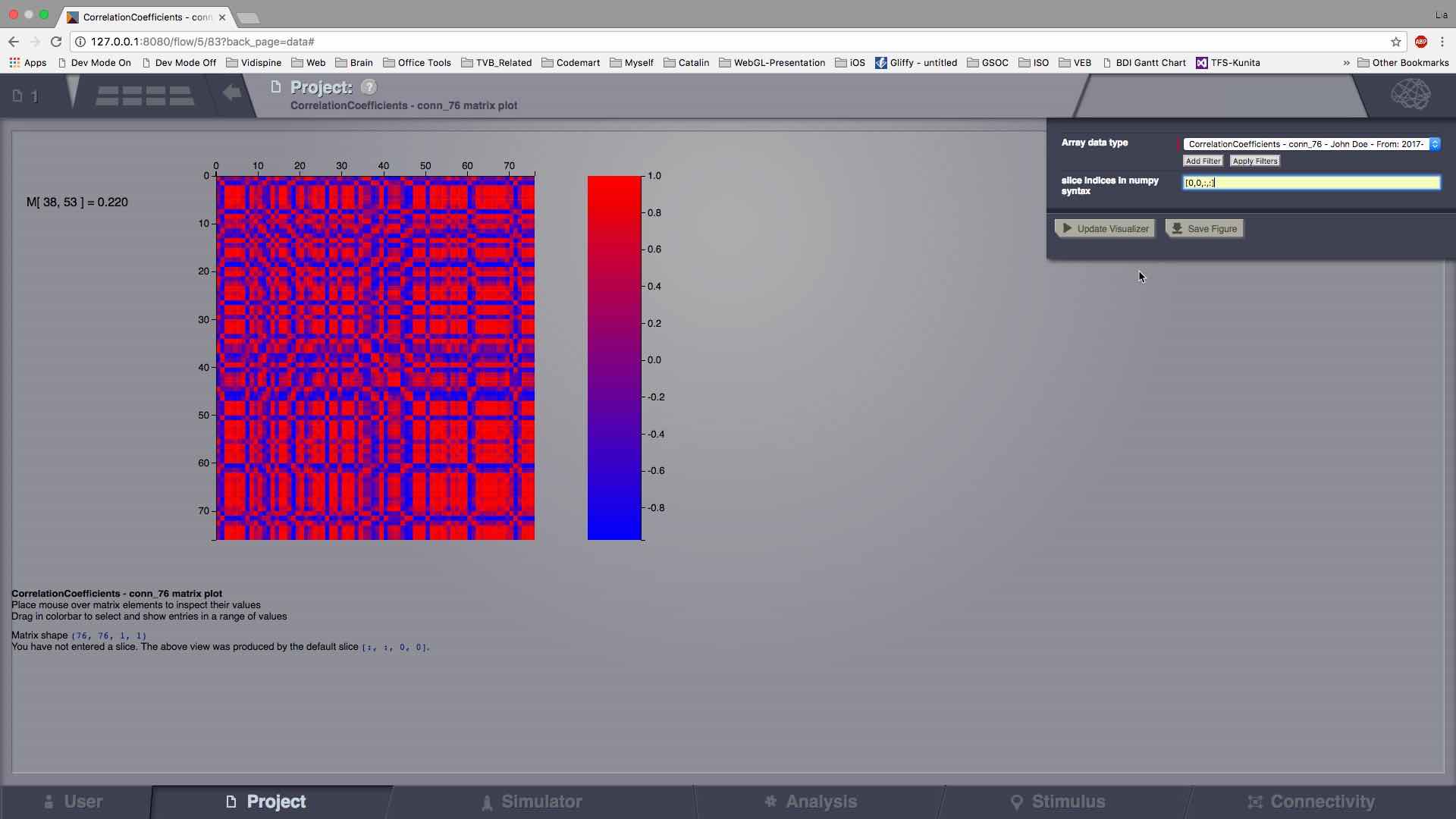
Preview for Matrix Visualizer¶
Connectivity Edge Bundle Visualizer¶
Shows structural connectivity coming in and out of a brain region by highlighting paths to other regions.
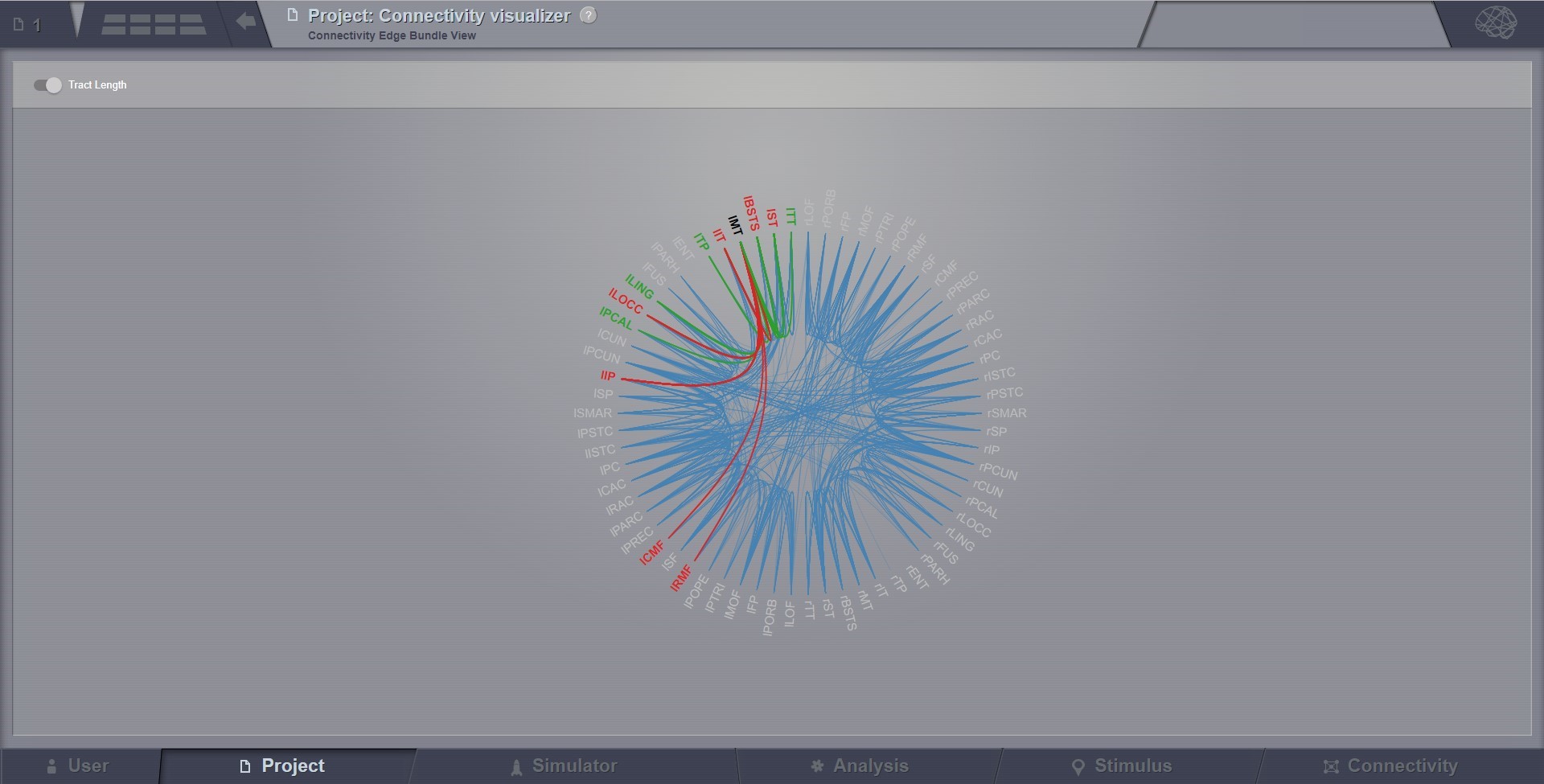
Preview for Connectivity Edge Bundle Visualizer¶
Pearson Edge Bundle Visualizer¶
Shows functional connectivity coming in and out of a brain region by highlighting paths to other regions.
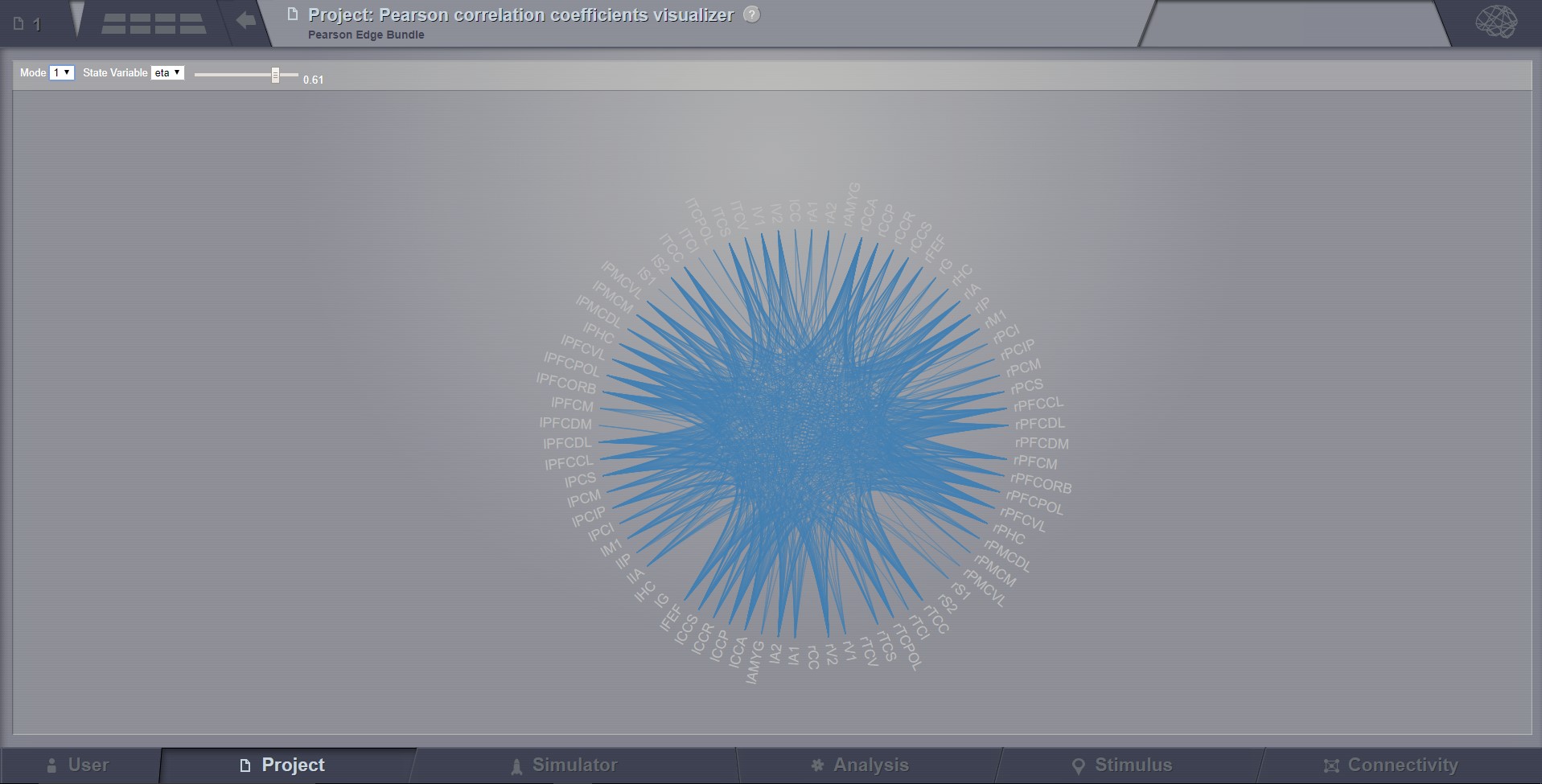
Preview for Pearson Edge Bundle Visualizer¶